Comparative physiological, biochemical, metabolomic, and transcriptomic analyses reveal the formation mechanism of heartwood for Acacia melanoxylon
- PMID: 38644502
- PMCID: PMC11034122
- DOI: 10.1186/s12870-024-04884-1
Comparative physiological, biochemical, metabolomic, and transcriptomic analyses reveal the formation mechanism of heartwood for Acacia melanoxylon
Abstract
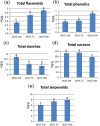
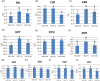
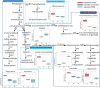
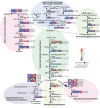
Acacia melanoxylon is well known as a valuable commercial tree species owing to its high-quality heartwood (HW) products. However, the metabolism and regulatory mechanism of heartwood during wood development remain largely unclear. In this study, both microscopic observation and content determination proved that total amount of starches decreased and phenolics and flavonoids increased gradually from sapwood (SW) to HW. We also obtained the metabolite profiles of 10 metabolites related to phenolics and flavonoids during HW formation by metabolomics. Additionally, we collected a comprehensive overview of genes associated with the biosynthesis of sugars, terpenoids, phenolics, and flavonoids using RNA-seq. A total of ninety-one genes related to HW formation were identified. The transcripts related to plant hormones, programmed cell death (PCD), and dehydration were increased in transition zone (TZ) than in SW. The results of RT-PCR showed that the relative expression level of genes and transcription factors was also high in the TZ, regardless of the horizontal or vertical direction of the trunk. Therefore, the HW formation took place in the TZ for A. melanoxylon from molecular level, and potentially connected to plant hormones, PCD, and cell dehydration. Besides, the increased expression of sugar and terpenoid biosynthesis-related genes in TZ further confirmed the close connection between terpenoid biosynthesis and carbohydrate metabolites of A. melanoxylon. Furthermore, the integrated analysis of metabolism data and RNA-seq data showed the key transcription factors (TFs) regulating flavonoids and phenolics accumulation in HW, including negative correlation TFs (WRKY, MYB) and positive correlation TFs (AP2, bZIP, CBF, PB1, and TCP). And, the genes and metabolites from phenylpropanoid and flavonoid metabolism and biosynthesis were up-regulated and largely accumulated in TZ and HW, respectively. The findings of this research provide a basis for comprehending the buildup of metabolites and the molecular regulatory processes of HW formation in A. melanoxylon.
Keywords: Acacia melanoxylon; Flavonoids; Heartwood formation; Metabolomic; Phenolics; Terpenoids; Transcriptomic.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Physiological, Biochemical, and Molecular Analyses Reveal Dark Heartwood Formation Mechanism in Acacia melanoxylon.Int J Mol Sci. 2024 May 2;25(9):4974. doi: 10.3390/ijms25094974. Int J Mol Sci. 2024. PMID: 38732191 Free PMC article.
-
Integrated Transcriptomic, Metabolomic, and Physiological Analyses Reveal New Insights into Fragrance Formation in the Heartwood of Phoebe hui.Int J Mol Sci. 2022 Nov 14;23(22):14044. doi: 10.3390/ijms232214044. Int J Mol Sci. 2022. PMID: 36430522 Free PMC article.
-
Comparative Metabolomics and Transcriptome Analysis Reveal the Fragrance-Related Metabolite Formation in Phoebe zhennan Wood.Molecules. 2023 Oct 12;28(20):7047. doi: 10.3390/molecules28207047. Molecules. 2023. PMID: 37894523 Free PMC article.
-
An extended model of heartwood secondary metabolism informed by functional genomics.Tree Physiol. 2018 Mar 1;38(3):311-319. doi: 10.1093/treephys/tpx070. Tree Physiol. 2018. PMID: 28633414 Review.
-
Role of bZIP transcription factors in the regulation of plant secondary metabolism.Planta. 2023 Jun 10;258(1):13. doi: 10.1007/s00425-023-04174-4. Planta. 2023. PMID: 37300575 Review.
Cited by
-
Advances in the Study of Heartwood Formation in Trees.Life (Basel). 2025 Jan 14;15(1):93. doi: 10.3390/life15010093. Life (Basel). 2025. PMID: 39860033 Free PMC article. Review.
-
Physiological, Biochemical, and Molecular Analyses Reveal Dark Heartwood Formation Mechanism in Acacia melanoxylon.Int J Mol Sci. 2024 May 2;25(9):4974. doi: 10.3390/ijms25094974. Int J Mol Sci. 2024. PMID: 38732191 Free PMC article.
-
CitGATA7 interact with histone acetyltransferase CitHAG28 to promote citric acid degradation by regulating the glutamine synthetase pathway in citrus.Mol Hortic. 2025 Feb 1;5(1):8. doi: 10.1186/s43897-024-00126-y. Mol Hortic. 2025. PMID: 39891226 Free PMC article.
-
Integrated transcriptomic and proteomic analyses provide insights into the biosynthesis of Lycii fructus polysaccharides from different cultivation regions.Food Chem (Oxf). 2024 Dec 5;10:100232. doi: 10.1016/j.fochms.2024.100232. eCollection 2025 Jun. Food Chem (Oxf). 2024. PMID: 39741492 Free PMC article.
References
-
- Nicholas I. Understanding blackwood (Acacia melanoxylon) markets; an opportunity for improving blackwood plantation returns. new Z J Forestry. 2007;52(3):17–20.
-
- Machado JS, Louzada JL, Santos AJA, Nunes L, Anjos O, Rodrigues J, Simoes RMS, Pereira H. Variation of Wood Density and Mechanical Properties of Blackwood (Acacia melanoxylon R. Br) Mater Design. 2014;56(apr):975–80. doi: 10.1016/j.matdes.2013.12.016. - DOI
-
- Spicer R. Senescence in secondary xylem: heartwood formation as an active Developmental Program. Vascular Transp Plants 2005:457–75.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

