Identifying MS4A6A+ macrophages as potential contributors to the pathogenesis of nonalcoholic fatty liver disease, periodontitis, and type 2 diabetes mellitus
- PMID: 38644829
- PMCID: PMC11033123
- DOI: 10.1016/j.heliyon.2024.e29340
Identifying MS4A6A+ macrophages as potential contributors to the pathogenesis of nonalcoholic fatty liver disease, periodontitis, and type 2 diabetes mellitus
Abstract
Purpose: Concrete epidemiological evidence has suggested the mutually-contributing effect respectively between nonalcoholic fatty liver disease (NAFLD), type 2 diabetes mellitus (T2DM), and periodontitis (PD); however, their shared crosstalk mechanism remains an open issue.
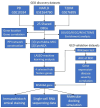
Method: The NAFLD, PD, and T2DM-related datasets were obtained from the NCBI GEO repository. Their common differentially expressed genes (DEGs) were identified and the functional enrichment analysis performed by the DAVID platform determined relevant biological processes and pathways. Then, the STRING database established a PPI network of such DEGs and topological analysis through Cytoscape 3.7.1 software along with the machine-learning analysis by the least absolute shrinkage and selection operator (LASSO) algorithm screened out hub characteristic genes. Their efficacy was validated by external datasets using the receiver operating characteristic (ROC) curve, and gene expression and location of the most robust one was determined using single-cell sequencing and immunohistochemical staining. Finally, the promising drugs were predicted through the CTD database, and the CB-DOCK 2 and Pymol platform mimicked molecular docking.
Result: Intersection of differentially expressed genes from three datasets identified 25 shared DEGs of the three diseases, which were enriched in MHC II-mediated antigen presenting process. PPI network and LASSO machine-learning analysis determined 4 feature genes, of which the MS4A6A gene mainly expressed by macrophages was the hub gene and key immune cell type. Molecular docking simulation chosen fenretinide as the most promising medicant for MS4A6A+ macrophages.
Conclusion: MS4A6A+ macrophages were suggested to be important immune-related mediators in the progression of NAFLD, PD, and T2DM pathologies.
Keywords: MS4A6A gene; Macrophages; Nonalcoholic fatty liver disease; Periodontitis; Type 2 diabetes mellitus.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






Similar articles
-
An integrated bioinformatics analysis to identify the shared biomarkers in patients with obstructive sleep apnea syndrome and nonalcoholic fatty liver disease.Front Genet. 2024 Jul 16;15:1356105. doi: 10.3389/fgene.2024.1356105. eCollection 2024. Front Genet. 2024. PMID: 39081807 Free PMC article.
-
Integrative analysis identifies oxidative stress biomarkers in non-alcoholic fatty liver disease via machine learning and weighted gene co-expression network analysis.Front Immunol. 2024 Feb 27;15:1335112. doi: 10.3389/fimmu.2024.1335112. eCollection 2024. Front Immunol. 2024. PMID: 38476236 Free PMC article.
-
Renal tubular gen e biomarkers identification based on immune infiltrates in focal segmental glomerulosclerosis.Ren Fail. 2022 Dec;44(1):966-986. doi: 10.1080/0886022X.2022.2081579. Ren Fail. 2022. PMID: 35713363 Free PMC article.
-
Discovering genetic linkage between periodontitis and type 1 diabetes: A bioinformatics study.Front Genet. 2023 Mar 27;14:1147819. doi: 10.3389/fgene.2023.1147819. eCollection 2023. Front Genet. 2023. PMID: 37051594 Free PMC article.
-
Identification of hub biomarkers of myocardial infarction by single-cell sequencing, bioinformatics, and machine learning.Front Cardiovasc Med. 2022 Jul 25;9:939972. doi: 10.3389/fcvm.2022.939972. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 35958412 Free PMC article.
References
-
- Slots J. Periodontitis: facts, fallacies and the future. Periodontol. 2000. 2017;75(1):7–23. - PubMed
-
- Chen M.X., et al. Global, regional, and national burden of severe periodontitis, 1990-2019: an analysis of the Global Burden of Disease Study 2019. J. Clin. Periodontol. 2021;48(9):1165–1188. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

