Deciphering the role of non-coding RNAs involved in sorafenib resistance
- PMID: 38644890
- PMCID: PMC11031791
- DOI: 10.1016/j.heliyon.2024.e29374
Deciphering the role of non-coding RNAs involved in sorafenib resistance
Abstract
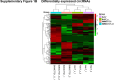
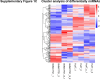
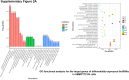
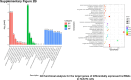
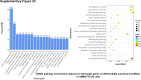
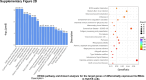
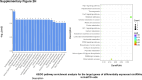
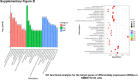
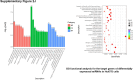
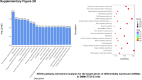
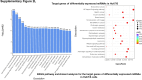
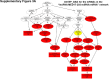
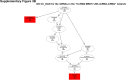
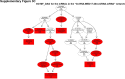
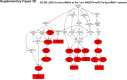
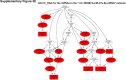
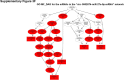
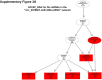
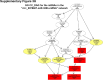
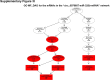
Sorafenib is an important treatment strategy for advanced hepatocellular carcinoma (HCC). Unfortunately, drug resistance has become a major obstacle in sorafenib application. In this study, whole transcriptome sequencing (WTS) was conducted to compare the paired differences between non-coding RNAs (ncRNAs), including long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), microRNAs (miRNAs), and mRNAs, in sorafenib-resistant and parental cells. The overlap of differentially expressed ncRNAs (DENs) between the SMMC7721/S and Huh7/S cells and their parental cells was determined. 2 upregulated and 3 downregulated lncRNAs, 2 upregulated and 1 downregulated circRNAs, as well as 10 upregulated and 2 downregulated miRNAs, in both SMMC7721/S and Huh7/S cells, attracted more attention. The target genes of these DENs were then identified as the overlaps between the differentially expressed mRNAs achieved using the WTS analysis and the predicted genes of DENs obtained using the "co-localization" or "co-expression," miRanda, and RNAhybrid analysis. Consequently, the potential regulatory network between overlapping DENs and their target genes in both SMMC7721/S and Huh7/S cells was explored. The "lncRNA-miRNA-mRNA" and "circRNA-miRNA-mRNA" networks were constructed based on the competitive endogenous RNA (ceRNA) theory using the Cytoscape software. In particular, lncRNA MED17-203-miRNA (miR-193a-5p, miR-197-3p, miR-27a-5p, miR-320b, miR-767-3p, miR-767-5p, miR-92a-3p, let-7c-5p)-mRNA," "circ_0002874-miR-27a-5p-mRNA" and "circ_0078607-miR-320b-mRNA" networks were first introduced in sorafenib-resistant HCC. Furthermore, these networks were most probably connected to the process of metabolic reprogramming, where the activation of the PPAR, HIF-1, Hippo, and TGF-β signaling pathways is governed. Alternatively, the network "circ_0002874-miR-27a-5p-mRNA" was also involved in the regulation of the activation of TGF-β signaling pathways, thus advancing Epithelial-mesenchymal transition (EMT). These findings provide a theoretical basis for exploring the mechanisms underlying sorafenib resistance mediated by metabolic reprogramming and EMT in HCC.
Keywords: Circular RNAs; Hepatocellular carcinoma; Long non-coding RNAs; MicroRNAs; Non-coding RNAs; Sorafenib.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






Similar articles
-
Circ_0001944 Targets the miR-1292-5p/FBLN2 Axis to Facilitate Sorafenib Resistance in Hepatocellular Carcinoma by Impeding Ferroptosis.Immunotargets Ther. 2024 Nov 26;13:643-659. doi: 10.2147/ITT.S463556. eCollection 2024. Immunotargets Ther. 2024. PMID: 39624827 Free PMC article.
-
[Non-coding RNAs expression profile of adjacent and distant liver tissues of hepatic cystic echinococcosis lesions].Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2025 May 9;37(2):152-162. doi: 10.16250/j.32.1915.2024216. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2025. PMID: 40425498 Chinese.
-
Identification of the circRNA-miRNA-mRNA Regulatory Network in Pterygium-Associated Conjunctival Epithelium.Biomed Res Int. 2022 Nov 8;2022:2673890. doi: 10.1155/2022/2673890. eCollection 2022. Biomed Res Int. 2022. PMID: 36398070 Free PMC article.
-
Epigenetic Associations between lncRNA/circRNA and miRNA in Hepatocellular Carcinoma.Cancers (Basel). 2020 Sep 14;12(9):2622. doi: 10.3390/cancers12092622. Cancers (Basel). 2020. PMID: 32937886 Free PMC article. Review.
-
Differentially expressed non-coding RNAs and their regulatory networks in liver cancer.Heliyon. 2023 Aug 19;9(9):e19223. doi: 10.1016/j.heliyon.2023.e19223. eCollection 2023 Sep. Heliyon. 2023. PMID: 37662778 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

