This is a preprint.
Muscle-Specific Pyruvate Kinase Isoforms, Pkm1 and Pkm2, Regulate Mammalian SWI/SNF Proteins and Histone 3 Phosphorylation During Myoblast Differentiation
- PMID: 38645038
- PMCID: PMC11030359
- DOI: 10.1101/2024.04.10.588959
Muscle-Specific Pyruvate Kinase Isoforms, Pkm1 and Pkm2, Regulate Mammalian SWI/SNF Proteins and Histone 3 Phosphorylation During Myoblast Differentiation
Update in
-
Muscle-specific pyruvate kinase isoforms, PKM1 and PKM2, regulate mammalian SWI/SNF proteins and histone 3 phosphorylation during myoblast differentiation.FASEB J. 2024 Jun 15;38(11):e23702. doi: 10.1096/fj.202400784R. FASEB J. 2024. PMID: 38837439 Free PMC article.
Abstract
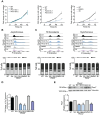
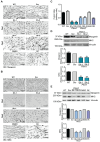
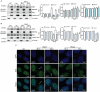
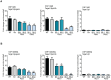
Pyruvate kinase is a glycolytic enzyme that converts phosphoenolpyruvate and ADP into pyruvate and ATP. There are two genes that encode pyruvate kinase in vertebrates; Pkm and Pkl encode muscle- and liver/erythrocyte-specific forms, respectively. Each gene encodes two isoenzymes due to alternative splicing. Both muscle-specific enzymes, Pkm1 and Pkm2, function in glycolysis, but Pkm2 also has been implicated in gene regulation due to its ability to phosphorylate histone 3 threonine 11 (H3T11) in cancer cells. Here, we examined the roles of Pkm1 and Pkm2 during myoblast differentiation. RNA-seq analysis revealed that Pkm2 promotes the expression of Dpf2/Baf45d and Baf250a/Arid1A. Dpf2 and Baf250a are subunits that identify a specific sub-family of the mammalian SWI/SNF (mSWI/SNF) of chromatin remodeling enzymes that is required for activation of myogenic gene expression during differentiation. Pkm2 also mediated the incorporation of Dpf2 and Baf250a into the regulatory sequences controlling myogenic gene expression. Pkm1 did not affect expression but was required for nuclear localization of Dpf2. Additionally, Pkm2 was required not only for the incorporation of phosphorylated H3T11 in myogenic promoters, but also for the incorporation of phosphorylated H3T6 and H3T45 at myogenic promoters via regulation of AKT and protein kinase C isoforms that phosphorylate those amino acids. Our results identify multiple unique roles for Pkm2 and a novel function for Pkm1 in gene expression and chromatin regulation during myoblast differentiation.
Keywords: H3 phosphorylation; Pyruvate kinase; SWI/SNF; chromatin remodeling enzymes; gene regulation; myoblast differentiation.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT The authors declare no conflicts of interest.
Figures




Similar articles
-
Muscle-specific pyruvate kinase isoforms, PKM1 and PKM2, regulate mammalian SWI/SNF proteins and histone 3 phosphorylation during myoblast differentiation.FASEB J. 2024 Jun 15;38(11):e23702. doi: 10.1096/fj.202400784R. FASEB J. 2024. PMID: 38837439 Free PMC article.
-
Differential Contributions of mSWI/SNF Chromatin Remodeler Sub-Families to Myoblast Differentiation.Int J Mol Sci. 2023 Jul 9;24(14):11256. doi: 10.3390/ijms241411256. Int J Mol Sci. 2023. PMID: 37511016 Free PMC article.
-
Glucose Catabolism in Liver Tumors Induced by c-MYC Can Be Sustained by Various PKM1/PKM2 Ratios and Pyruvate Kinase Activities.Cancer Res. 2017 Aug 15;77(16):4355-4364. doi: 10.1158/0008-5472.CAN-17-0498. Epub 2017 Jun 19. Cancer Res. 2017. PMID: 28630053 Free PMC article.
-
Regulation of the Mammalian SWI/SNF Family of Chromatin Remodeling Enzymes by Phosphorylation during Myogenesis.Biology (Basel). 2020 Jul 3;9(7):152. doi: 10.3390/biology9070152. Biology (Basel). 2020. PMID: 32635263 Free PMC article. Review.
-
Pyruvate Kinase M2 and Cancer: The Role of PKM2 in Promoting Tumorigenesis.Front Oncol. 2020 Mar 2;10:159. doi: 10.3389/fonc.2020.00159. eCollection 2020. Front Oncol. 2020. PMID: 32195169 Free PMC article. Review.
References
-
- Chal J., and Pourquie O. (2017) Making muscle: skeletal myogenesis in vivo and in vitro. Development 144, 2104–2122 - PubMed
-
- Stern H. M., Brown A. M., and Hauschka S. D. (1995) Myogenesis in paraxial mesoderm: preferential induction by dorsal neural tube and by cells expressing Wnt-1. Development 121, 3675–3686 - PubMed
-
- Tajbakhsh S., Borello U., Vivarelli E., Kelly R., Papkoff J., Duprez D., Buckingham M., and Cossu G. (1998) Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development 125, 4155–4162 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
