This is a preprint.
A framework for quality control in quantitative proteomics
- PMID: 38645098
- PMCID: PMC11030400
- DOI: 10.1101/2024.04.12.589318
A framework for quality control in quantitative proteomics
Update in
-
A Framework for Quality Control in Quantitative Proteomics.J Proteome Res. 2024 Oct 4;23(10):4392-4408. doi: 10.1021/acs.jproteome.4c00363. Epub 2024 Sep 9. J Proteome Res. 2024. PMID: 39248652 Free PMC article.
Abstract
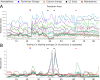
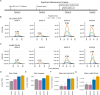
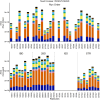
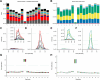
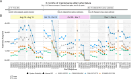
A thorough evaluation of the quality, reproducibility, and variability of bottom-up proteomics data is necessary at every stage of a workflow from planning to analysis. We share vignettes applying adaptable quality control (QC) measures to assess sample preparation, system function, and quantitative analysis. System suitability samples are repeatedly measured longitudinally with targeted methods, and we share examples where they are used on three instrument platforms to identify severe system failures and track function over months to years. Internal QCs incorporated at protein and peptide-level allow our team to assess sample preparation issues and to differentiate system failures from sample-specific issues. External QC samples prepared alongside our experimental samples are used to verify the consistency and quantitative potential of our results during batch correction and normalization before assessing biological phenotypes. We combine these controls with rapid analysis (Skyline), longitudinal QC metrics (AutoQC), and server-based data deposition (PanoramaWeb). We propose that this integrated approach to QC is a useful starting point for groups to facilitate rapid quality control assessment to ensure that valuable instrument time is used to collect the best quality data possible. Data are available on Panorama Public and on ProteomeXchange under the identifier PXD051318.
Keywords: DDA; DIA; PRM; liquid chromatography; mass spectrometry; proteomics; quality control; quantitative results; sample preparation; system suitability.
Conflict of interest statement
Conflict of interest statement The authors declare the following competing financial interest(s): The MacCoss Lab at the University of Washington has a sponsored research agreement with Thermo Fisher Scientific, the manufacturer of the instrumentation used in this research. M.J.M. is a paid consultant for Thermo Fisher Scientific. J.D.C. is an employee of Thermo Fisher Scientific
Figures


References
-
- Piehowski P. D.; Petyuk V. A.; Orton D. J.; Xie F.; Moore R. J.; Ramirez-restrepo M.; Engel A.; Lieberman A. P.; Albin R. L.; Camp D. G.; Smith R. D.; Myers A. J. Sources of Technical Variability in Quantitative LC − MS Proteomics: Human Brain Tissue Sample Analysis. J. Proteome Res. 2013, 12, 2128–2137. 10.1021/pr301146m. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
