This is a preprint.
Classification and functional characterization of regulators of intracellular STING trafficking identified by genome-wide optical pooled screening
- PMID: 38645119
- PMCID: PMC11030420
- DOI: 10.1101/2024.04.07.588166
Classification and functional characterization of regulators of intracellular STING trafficking identified by genome-wide optical pooled screening
Update in
-
Classification and functional characterization of regulators of intracellular STING trafficking identified by genome-wide optical pooled screening.Cell Syst. 2024 Dec 18;15(12):1264-1277.e8. doi: 10.1016/j.cels.2024.11.004. Epub 2024 Dec 9. Cell Syst. 2024. PMID: 39657680
Abstract
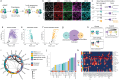
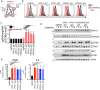
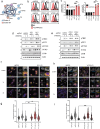
STING is an innate immune sensor that traffics across many cellular compartments to carry out its function of detecting cyclic di-nucleotides and triggering defense processes. Mutations in factors that regulate this process are often linked to STING-dependent human inflammatory disorders. To systematically identify factors involved in STING trafficking, we performed a genome-wide optical pooled screen and examined the impact of genetic perturbations on intracellular STING localization. Based on subcellular imaging of STING protein and trafficking markers in 45 million cells perturbed with sgRNAs, we defined 464 clusters of gene perturbations with similar cellular phenotypes. A higher-dimensional focused optical pooled screen on 262 perturbed genes which assayed 11 imaging channels identified 73 finer phenotypic clusters. In a cluster containing USE1, a protein that mediates Golgi to ER transport, we found a gene of unknown function, C19orf25. Consistent with the known role of USE1, loss of C19orf25 enhanced STING signaling. Other clusters contained subunits of the HOPS, GARP and RIC1-RGP1 complexes. We show that HOPS deficiency delayed STING degradation and consequently increased signaling. Similarly, GARP/RIC1-RGP1 loss increased STING signaling by delaying STING exit from the Golgi. Our findings demonstrate that genome-wide genotype-phenotype maps based on high-content cell imaging outperform other screening approaches, and provide a community resource for mining for factors that impact STING trafficking as well as other cellular processes observable in our dataset.
Conflict of interest statement
Competing Interests P.C.B. is a consultant to or holds equity in 10X Genomics, General Automation Lab Technologies/Isolation Bio, Celsius Therapeutics, Next Gen Diagnostics, Cache DNA, Concerto Biosciences, Stately, Ramona Optics, Bifrost Biosystems, and Amber Bio. His laboratory has received research funding from Calico Life Sciences, Merck, and Genentech for work related to genetic screening. N.H. holds equity in and advises Danger Bio/Related Sciences, is on the scientific advisory board of Repertoire Immune Medicines and CytoReason, owns equity and has licensed patents to BioNtech, and receives research funding from Bristol Myers Squibb and Calico Life Sciences. The Broad Institute and MIT may seek to commercialize aspects of this work, and related applications for intellectual property have been filed. R.J.C. is an employee of Flagship Pioneering.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
