Analysis of weighted gene co-expression networks and clinical validation identify hub genes and immune cell infiltration in the endometrial cells of patients with recurrent implantation failure
- PMID: 38645487
- PMCID: PMC11026622
- DOI: 10.3389/fgene.2024.1292757
Analysis of weighted gene co-expression networks and clinical validation identify hub genes and immune cell infiltration in the endometrial cells of patients with recurrent implantation failure
Abstract
Background: About 10% of individuals undergoing in vitro fertilization encounter recurrent implantation failure (RIF), which represents a worldwide social and economic concern. Nevertheless, the critical genes and genetic mechanisms underlying RIF are largely unknown.
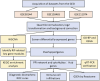
Methods: We first obtained three comprehensive microarray datasets "GSE58144, GSE103465 and GSE111974". The differentially expressed genes (DEGs) evaluation, enrichment analysis, as well as efficient weighted gene co-expression network analysis (WGCNA), were employed for distinguishing RIF-linked hub genes, which were tested by RT-qPCR in our 30 independent samples. Next, we studied the topography of infiltration of 22 immune cell subpopulations and the association between hub genes and immune cells in RIF using the CIBERSORT algorithm. Finally, a novel ridge plot was utilized to exhibit the potential function of core genes.
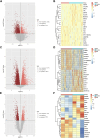
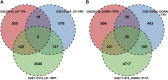
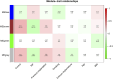
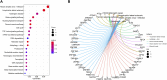
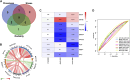
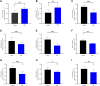
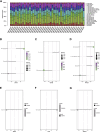
Results: The enrichment of GO/KEGG pathways reveals that Herpes simplex virus 1 infection and Salmonella infection may have an important role in RIF. After WGCNA, the intersected genes with the previous DEGs were obtained using both variance and association. Notably, the subsequent nine hub genes were finally selected: ACTL6A, BECN1, SNRPD1, POLR1B, GSK3B, PPP2CA, RBBP7, PLK4, and RFC4, based on the PPI network and three different algorithms, whose expression patterns were also verified by RT-qPCR. With in-depth analysis, we speculated that key genes mentioned above might be involved in the RIF through disturbing endometrial microflora homeostasis, impairing autophagy, and inhibiting the proliferation of endometrium. Furthermore, the current study revealed the aberrant immune infiltration patterns and emphasized that uterine NK cells (uNK) and CD4+ T cells were substantially altered in RIF endometrium. Finally, the ridge plot displayed a clear and crucial association between hub genes and other genes and key pathways.
Conclusion: We first utilized WGCNA to identify the most potential nine hub genes which might be associated with RIF. Meanwhile, this study offers insights into the landscape of immune infiltration status to reveal the underlying immune pathogenesis of RIF. This may be a direction for the next study of RIF etiology. Further studies would be required to investigate the involved mechanisms.
Keywords: bioinformatics; hub genes; immune cell infiltration; infertility; recurrent implantation failure.
Copyright © 2024 Liu, Lai, Qu, Liu, Zhang, Zhao, He, Sun and Bao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Ahmadi M., Pashangzadeh S., Moraghebi M., Sabetian S., Shekari M., Eini F., et al. (2022). Construction of circRNA-miRNA-mRNA network in the pathogenesis of recurrent implantation failure using integrated bioinformatics study. J. Cell Mol. Med. 26 (6), 1853–1864. 10.1111/jcmm.16586 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

