Changes in total charge on spike protein of SARS-CoV-2 in emerging lineages
- PMID: 38645718
- PMCID: PMC11031363
- DOI: 10.1093/bioadv/vbae053
Changes in total charge on spike protein of SARS-CoV-2 in emerging lineages
Abstract
Motivation: Charged amino acid residues on the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been shown to influence its binding to different cell surface receptors, its non-specific electrostatic interactions with the environment, and its structural stability and conformation. It is therefore important to obtain a good understanding of amino acid mutations that affect the total charge on the spike protein which have arisen across different SARS-CoV-2 lineages during the course of the virus' evolution.
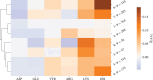
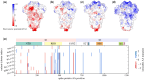
Results: We analyse the change in the number of ionizable amino acids and the corresponding total charge on the spike proteins of almost 2200 SARS-CoV-2 lineages that have emerged over the span of the pandemic. Our results show that the previously observed trend toward an increase in the positive charge on the spike protein of SARS-CoV-2 variants of concern has essentially stopped with the emergence of the early omicron variants. Furthermore, recently emerged lineages show a greater diversity in terms of their composition of ionizable amino acids. We also demonstrate that the patterns of change in the number of ionizable amino acids on the spike protein are characteristic of related lineages within the broader clade division of the SARS-CoV-2 phylogenetic tree. Due to the ubiquity of electrostatic interactions in the biological environment, our findings are relevant for a broad range of studies dealing with the structural stability of SARS-CoV-2 and its interactions with the environment.
Availability and implementation: The data underlying the article are available in the Supplementary material.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
No competing interest is declared.
Figures

Similar articles
-
Evolutionary changes in the number of dissociable amino acids on spike proteins and nucleoproteins of SARS-CoV-2 variants.Virus Evol. 2023 Jun 29;9(2):vead040. doi: 10.1093/ve/vead040. eCollection 2023. Virus Evol. 2023. PMID: 37583936 Free PMC article.
-
SARS-CoV-2 nonstructural protein 6 from Alpha to Omicron: evolution of a transmembrane protein.mBio. 2023 Aug 31;14(4):e0068823. doi: 10.1128/mbio.00688-23. Epub 2023 Jul 21. mBio. 2023. PMID: 37477426 Free PMC article.
-
Dissecting Naturally Arising Amino Acid Substitutions at Position L452 of SARS-CoV-2 Spike.J Virol. 2022 Oct 26;96(20):e0116222. doi: 10.1128/jvi.01162-22. Epub 2022 Oct 10. J Virol. 2022. PMID: 36214577 Free PMC article.
-
Severe Acute Respiratory Syndrome Coronavirus 2 Variants of Concern: A Perspective for Emerging More Transmissible and Vaccine-Resistant Strains.Viruses. 2022 Apr 16;14(4):827. doi: 10.3390/v14040827. Viruses. 2022. PMID: 35458557 Free PMC article. Review.
-
The effects of amino acid substitution of spike protein and genomic recombination on the evolution of SARS-CoV-2.Front Microbiol. 2023 Jul 25;14:1228128. doi: 10.3389/fmicb.2023.1228128. eCollection 2023. Front Microbiol. 2023. PMID: 37560529 Free PMC article. Review.
Cited by
-
Biophysics of SARS-CoV-2 spike protein's receptor-binding domain interaction with ACE2 and neutralizing antibodies: from computation to functional insights.Biophys Rev. 2025 Mar 8;17(2):309-333. doi: 10.1007/s12551-025-01276-z. eCollection 2025 Apr. Biophys Rev. 2025. PMID: 40376405 Review.
-
Quantitative Characterization and Prediction of the Binding Determinants and Immune Escape Hotspots for Groups of Broadly Neutralizing Antibodies Against Omicron Variants: Atomistic Modeling of the SARS-CoV-2 Spike Complexes with Antibodies.Biomolecules. 2025 Feb 8;15(2):249. doi: 10.3390/biom15020249. Biomolecules. 2025. PMID: 40001552 Free PMC article.
-
Semi-Covariance Coefficient Analysis of Spike Proteins from SARS-CoV-2 and Its Variants Omicron, BA.5, EG.5, and JN.1 for Viral Infectivity, Virulence and Immune Escape.Viruses. 2024 Jul 25;16(8):1192. doi: 10.3390/v16081192. Viruses. 2024. PMID: 39205166 Free PMC article.
-
Conformational and Stability Analysis of SARS-CoV-2 Spike Protein Variants by Molecular Simulation.Pathogens. 2025 Mar 12;14(3):274. doi: 10.3390/pathogens14030274. Pathogens. 2025. PMID: 40137759 Free PMC article.
-
Large-Scale FMO-MP2 Calculations of the Spike Protein Droplet Model.J Comput Chem. 2025 Feb 5;46(4):e70052. doi: 10.1002/jcc.70052. J Comput Chem. 2025. PMID: 39894970 Free PMC article.
References
-
- Aksamentov I, Roemer C, Hodcroft EB. et al. Nextclade: clade assignment, mutation calling and quality control for viral genomes. JOSS 2021;6:3773.
LinkOut - more resources
Full Text Sources
Miscellaneous

