pH-responsive i-motif-conjugated nanoparticles for MRI analysis
- PMID: 38646186
- PMCID: PMC11025034
- DOI: 10.1039/d3sd00285c
pH-responsive i-motif-conjugated nanoparticles for MRI analysis
Abstract
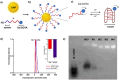
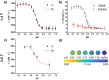
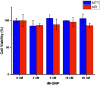
Gadolinium (Gd)-based contrast agents (CAs) are widely used to enhance anatomical details in magnetic resonance imaging (MRI). Significant research has expanded the field of CAs into bioresponsive CAs by modulating the signal to image and monitor biochemical processes, such as pH. In this work, we introduce the modular, dynamic actuation mechanism of DNA-based nanostructures as a new way to modulate the MRI signal based on the rotational correlation time, τR. We combined a pH-responsive oligonucleotide (i-motif) and a clinical standard CA (Gd-DOTA) to develop a pH-responsive MRI CA. The i-motif folds into a quadruplex under acidic conditions and was incorporated onto gold nanoparticles (iM-GNP) to achieve increased relaxivity, r1, compared to the unbound i-motif. In vitro, iM-GNP resulted in a significant increase in r1 over a decreasing pH range (7.5-4.5) with a calculated pKa = 5.88 ± 0.01 and a 16.7% change per 0.1 pH unit. In comparison, a control CA with a non-responsive DNA strand (T33-GNP) did not show a significant change in r1 over the same pH range. The iM-GNP was further evaluated in 20% human serum and demonstrated a 28.14 ± 11.2% increase in signal from neutral pH to acidic pH. This approach paves a path for novel programmable, dynamic DNA-based complexes for τR-modulated bioresponsive MRI CAs.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures

References
-
- Moats R. A. Fraser S. E. Meade T. J. Angew. Chem., Int. Ed. Engl. 1997;36:726–728. doi: 10.1002/anie.199707261. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
