MHCII-peptide presentation: an assessment of the state-of-the-art prediction methods
- PMID: 38646540
- PMCID: PMC11027168
- DOI: 10.3389/fimmu.2024.1293706
MHCII-peptide presentation: an assessment of the state-of-the-art prediction methods
Abstract
Major histocompatibility complex Class II (MHCII) proteins initiate and regulate immune responses by presentation of antigenic peptides to CD4+ T-cells and self-restriction. The interactions between MHCII and peptides determine the specificity of the immune response and are crucial in immunotherapy and cancer vaccine design. With the ever-increasing amount of MHCII-peptide binding data available, many computational approaches have been developed for MHCII-peptide interaction prediction over the last decade. There is thus an urgent need to provide an up-to-date overview and assessment of these newly developed computational methods. To benchmark the prediction performance of these methods, we constructed an independent dataset containing binding and non-binding peptides to 20 human MHCII protein allotypes from the Immune Epitope Database, covering DP, DR and DQ alleles. After collecting 11 known predictors up to January 2022, we evaluated those available through a webserver or standalone packages on this independent dataset. The benchmarking results show that MixMHC2pred and NetMHCIIpan-4.1 achieve the best performance among all predictors. In general, newly developed methods perform better than older ones due to the rapid expansion of data on which they are trained and the development of deep learning algorithms. Our manuscript not only draws a full picture of the state-of-art of MHCII-peptide binding prediction, but also guides researchers in the choice among the different predictors. More importantly, it will inspire biomedical researchers in both academia and industry for the future developments in this field.
Keywords: MHCII; bioinformatics; immunology; machine learning; peptide binding prediction; webserver.
Copyright © 2024 Yang, Wei, Cia, Song, Pucci, Rooman, Xue and Hou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
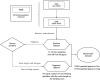
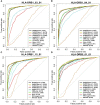
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

