Two sequential gene expression programs bridged by cell division support long-distance collective cell migration
- PMID: 38646822
- PMCID: PMC11165717
- DOI: 10.1242/dev.202262
Two sequential gene expression programs bridged by cell division support long-distance collective cell migration
Abstract
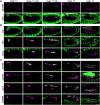
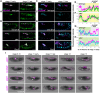
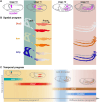
The precise assembly of tissues and organs relies on spatiotemporal regulation of gene expression to coordinate the collective behavior of cells. In Drosophila embryos, the midgut musculature is formed through collective migration of caudal visceral mesoderm (CVM) cells, but how gene expression changes as cells migrate is not well understood. Here, we have focused on ten genes expressed in the CVM and the cis-regulatory sequences controlling their expression. Although some genes are continuously expressed, others are expressed only early or late during migration. Late expression relates to cell cycle progression, as driving string/Cdc25 causes earlier division of CVM cells and accelerates the transition to late gene expression. In particular, we found that the cell cycle effector transcription factor E2F1 is a required input for the late gene CG5080. Furthermore, whereas late genes are broadly expressed in all CVM cells, early gene transcripts are polarized to the anterior or posterior ends of the migrating collective. We show this polarization requires transcription factors Snail, Zfh1 and Dorsocross. Collectively, these results identify two sequential gene expression programs bridged by cell division that support long-distance directional migration of CVM cells.
Keywords: Caudal visceral mesoderm; Cell cycle progression; Collective cell migration; Spatiotemporal gene expression; Transcription programs.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

