The discovery of novel noncoding RNAs in 50 bacterial genomes
- PMID: 38647067
- PMCID: PMC11109978
- DOI: 10.1093/nar/gkae248
The discovery of novel noncoding RNAs in 50 bacterial genomes
Abstract
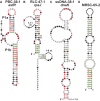
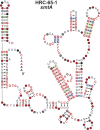
Structured noncoding RNAs (ncRNAs) contribute to many important cellular processes involving chemical catalysis, molecular recognition and gene regulation. Few ncRNA classes are broadly distributed among organisms from all three domains of life, but the list of rarer classes that exhibit surprisingly diverse functions is growing. We previously developed a computational pipeline that enables the near-comprehensive identification of structured ncRNAs expressed from individual bacterial genomes. The regions between protein coding genes are first sorted based on length and the fraction of guanosine and cytidine nucleotides. Long, GC-rich intergenic regions are then examined for sequence and structural similarity to other bacterial genomes. Herein, we describe the implementation of this pipeline on 50 bacterial genomes from varied phyla. More than 4700 candidate intergenic regions with the desired characteristics were identified, which yielded 44 novel riboswitch candidates and numerous other putative ncRNA motifs. Although experimental validation studies have yet to be conducted, this rate of riboswitch candidate discovery is consistent with predictions that many hundreds of novel riboswitch classes remain to be discovered among the bacterial species whose genomes have already been sequenced. Thus, many thousands of additional novel ncRNA classes likely remain to be discovered in the bacterial domain of life.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
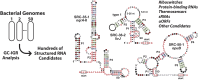



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

