The Subcortical Atlas of the Marmoset ("SAM") monkey based on high-resolution MRI and histology
- PMID: 38647221
- PMCID: PMC11494440
- DOI: 10.1093/cercor/bhae120
The Subcortical Atlas of the Marmoset ("SAM") monkey based on high-resolution MRI and histology
Abstract
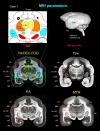
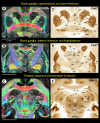
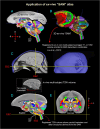
A comprehensive three-dimensional digital brain atlas of cortical and subcortical regions based on MRI and histology has a broad array of applications in anatomical, functional, and clinical studies. We first generated a Subcortical Atlas of the Marmoset, called the "SAM," from 251 delineated subcortical regions (e.g. thalamic subregions, etc.) derived from high-resolution Mean Apparent Propagator-MRI, T2W, and magnetization transfer ratio images ex vivo. We then confirmed the location and borders of these segmented regions in the MRI data using matched histological sections with multiple stains obtained from the same specimen. Finally, we estimated and confirmed the atlas-based areal boundaries of subcortical regions by registering this ex vivo atlas template to in vivo T1- or T2W MRI datasets of different age groups (single vs. multisubject population-based marmoset control adults) using a novel pipeline developed within Analysis of Functional NeuroImages software. Tracing and validating these important deep brain structures in 3D will improve neurosurgical planning, anatomical tract tracer injections, navigation of deep brain stimulation probes, functional MRI and brain connectivity studies, and our understanding of brain structure-function relationships. This new ex vivo template and atlas are available as volumes in standard NIFTI and GIFTI file formats and are intended for use as a reference standard for marmoset brain research.
Keywords: MAP-MRI; SAM; histology; marmoset monkey; subcortical atlas.
© The Author(s) 2024. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permission@oup.com.
Figures




Update of
-
The Subcortical Atlas of the Marmoset ("SAM") monkey based on high-resolution MRI and histology.bioRxiv [Preprint]. 2024 Jan 8:2024.01.06.574429. doi: 10.1101/2024.01.06.574429. bioRxiv. 2024. Update in: Cereb Cortex. 2024 Apr 1;34(4):bhae120. doi: 10.1093/cercor/bhae120. PMID: 38260391 Free PMC article. Updated. Preprint.
References
-
- Amaral DG, Bassett JL. Cholinergic innervation of the monkey amygdala: an immunohistochemical analysis with antisera to choline acetyltransferase. J Comp Neurol. 1989:281:337–361. - PubMed
-
- Avram AV, Guidon A, Song AW. Myelin water weighted diffusion tensor imaging. NeuroImage. 2010:53:132–138. - PubMed
Publication types
MeSH terms
Grants and funding
- Intramural Research Program of the National Institute of Neurological Disorders and Stroke
- Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development
- U01 EB026996/EB/NIBIB NIH HHS/United States
- 1U01EB026996-01/NIH BRAIN
- 309698-4.01-65310/CNRM Neuroradiology/Neuropathology Correlation/Integration Core
LinkOut - more resources
Full Text Sources
Medical

