Single-cell analysis revealed a potential role of T-cell exhaustion in colorectal cancer with liver metastasis
- PMID: 38647235
- PMCID: PMC11034372
- DOI: 10.1111/jcmm.18341
Single-cell analysis revealed a potential role of T-cell exhaustion in colorectal cancer with liver metastasis
Abstract
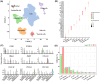
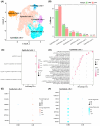
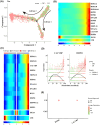
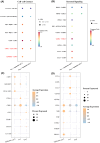
Liver metastasis (LM) is an important factor leading to colorectal cancer (CRC) mortality. However, the effect of T-cell exhaustion on LM in CRC is unclear. Single-cell sequencing data derived from the Gene Expression Omnibus database. Data were normalized using the Seurat package and subsequently clustered and annotated into different cell clusters. The differentiation trajectories of epithelial cells and T cells were characterized based on pseudo-time analysis. Single-sample gene set enrichment analysis (ssGSEA) was used to calculate enrichment scores for different cell clusters and to identify enriched biological pathways. Finally, cell communication analysis was performed. Nine cell subpopulations were identified from CRC samples with LM. The proportion of T cells increased in LM. T cells can be subdivided into NK/T cells, regulatory T cells (Treg) and exhausted T cells (Tex). In LM, cell adhesion and proliferation activity of Tex were promoted. Epithelial cells can be categorized into six subpopulations. The transformation of primary CRC into LM involved two evolutionary branches of Tex cells. Epithelial cells two were at the beginning of the trajectory in CRC but at the end of the trajectory in CRC with LM. The receptor ligands CEACAM5 and ADGRE5-CD55 played critical roles in the interactions between Tex and Treg cell-epithelial cell, which may promote the epithelial-mesenchymal transition process in CRC. Tex cells are able to promote the process of LM in CRC, which in turn promotes tumour development. This provides a new perspective on the treatment and diagnosis of CRC.
Keywords: exhausted T cells; liver metastasis; primary colorectal cancer; regulatory T cells; single‐cell sequencing.
© 2024 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflicts of interest regarding this manuscript.
Figures



References
-
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. - PubMed
-
- Ma S, Zhu X, Xin C, et al. RCN3 expression indicates prognosis in colorectal cancers. Oncologie. 2022;24(4):823‐833.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

