Lactobacillus Reuteri Vesicles Regulate Mitochondrial Function of Macrophages to Promote Mucosal and Cutaneous Wound Healing
- PMID: 38647360
- PMCID: PMC11199966
- DOI: 10.1002/advs.202309725
Lactobacillus Reuteri Vesicles Regulate Mitochondrial Function of Macrophages to Promote Mucosal and Cutaneous Wound Healing
Abstract
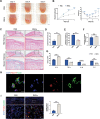
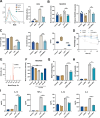
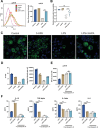
The interplay between bacteria and their host influences the homeostasis of the human immune microenvironment, and this reciprocal interaction also affects the process of tissue damage repair. A variety of immunomodulatory commensal bacteria reside in the body, capable of delivering membrane vesicles (MVs) to host cells to regulate the local immune microenvironment. This research revealed, for the initial time, the significant enhancement of mucosal and cutaneous wound healing by MVs secreted by the human commensal Lactobacillus reuteri (RMVs) through modulation of the inflammatory environment in wound tissue. Local administration of RMVs reduces the proportion of pro-inflammatory macrophages in inflamed tissues and mitigates the level of local inflammation, thereby facilitating the healing of oral mucosa and cutaneous wounds. The elevated oxidative stress levels in activated pro-inflammatory macrophages can be modulated by RMVs, resulting in phenotypic transformation of macrophages. Furthermore, 3-hydroxypropionaldehyde present in RMVs can decrease the mitochondrial permeability of macrophages and stabilize the mitochondrial membrane potential, thereby promoting the conversion of macrophages to an anti-inflammatory phenotype. This study pioneers the significance of commensal bacterial MVs in tissue injury repair and presents a novel concept for the repair of tissue damage.
Keywords: Lactobacillus reuteri DSM 20016; extracellular vesicles; macrophages; mitochondria; wound healing.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
