Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs
- PMID: 38647423
- PMCID: PMC11187927
- DOI: 10.1002/advs.202310120
Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs
Erratum in
-
Correction to "Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs".Adv Sci (Weinh). 2025 Feb;12(7):e2416290. doi: 10.1002/advs.202416290. Epub 2025 Jan 21. Adv Sci (Weinh). 2025. PMID: 39836607 Free PMC article. No abstract available.
Abstract
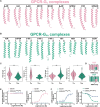
G-protein-coupled receptors (GPCRs) transmit downstream signals predominantly via G-protein pathways. However, the conformational basis of selective coupling of primary G-protein remains elusive. Histamine receptors H2R and H3R couple with Gs- or Gi-proteins respectively. Here, three cryo-EM structures of H2R-Gs and H3R-Gi complexes are presented at a global resolution of 2.6-2.7 Å. These structures reveal the unique binding pose for endogenous histamine in H3R, wherein the amino group interacts with E2065.46 of H3R instead of the conserved D1143.32 of other aminergic receptors. Furthermore, comparative analysis of the H2R-Gs and H3R-Gi complexes reveals that the structural geometry of TM5/TM6 determines the primary G-protein selectivity in histamine receptors. Machine learning (ML)-based structuromic profiling and functional analysis of class A GPCR-G-protein complexes illustrate that TM5 length, TM5 tilt, and TM6 outward movement are key determinants of the Gs and Gi/o selectivity among the whole Class A family. Collectively, the findings uncover the common structural geometry within class A GPCRs that determines the primary Gs- and Gi/o-coupling selectivity.
Keywords: G protein selectivity; GPCR; H2R; H3R; cryo‐EM structure; machine learning; signaling complex.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
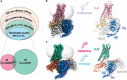


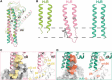


References
-
- a) Inoue A., Raimondi F., Kadji F. M. N., Singh G., Kishi T., Uwamizu A., Ono Y., Shinjo Y., Ishida S., Arang N., Kawakami K., Gutkind J. S., Aoki J., Russell R. B., Cell 2019, 177, 1933; - PMC - PubMed
- b) Sandhu M., Cho A., Ma N., Mukhaleva E., Namkung Y., Lee S., Ghosh S., Lee J. H., Gloriam D. E., Laporte S. A., Babu M. M., Vaidehi N., Nat. Commun. 2022, 13, 7428. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 92353303/National Natural Science Foundation of China
- 32141004/National Natural Science Foundation of China
- 32000845/National Natural Science Foundation of China
- U21A20418/National Natural Science Foundation of China
- 81973302/National Natural Science Foundation of China
- 62202426/National Natural Science Foundation of China
- 81825022/National Science Fund for Distinguished Young Scholars
- 2021C03039/Key R&D Projects of Zhejiang Province
- Fundamental Research Funds for the Central Universities
- 2019YFA050880/National Key Research and Development Program of China
- 2022ZD0205400/STI2030-Major Projects
- 2024C03147/"'Pioneer"' and "'Leading Goose"' R&D Program of Zhejiang
- 2020R01006/Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang
LinkOut - more resources
Full Text Sources
