A Nanopore Sequencing-based Pharmacogenomic Panel to Personalize Tuberculosis Drug Dosing
- PMID: 38647526
- PMCID: PMC11208962
- DOI: 10.1164/rccm.202309-1583OC
A Nanopore Sequencing-based Pharmacogenomic Panel to Personalize Tuberculosis Drug Dosing
Abstract
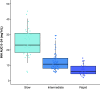
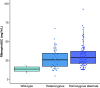
Rationale: Standardized dosing of antitubercular drugs leads to variable plasma drug levels, which are associated with adverse drug reactions, delayed treatment response, and relapse. Mutations in genes affecting drug metabolism explain considerable interindividual pharmacokinetic variability; however, pharmacogenomic assays that predict metabolism of antitubercular drugs have been lacking. Objectives: We sought to develop a Nanopore sequencing panel and validate its performance in patients with active tuberculosis (TB) to personalize treatment dosing. Methods: We developed a Nanopore sequencing panel targeting 15 SNPs in five genes affecting the metabolism of antitubercular drugs. For validation, we sequenced DNA samples (n = 48) from the 1,000 Genomes Project and compared the variant calling accuracy with that of Illumina genome sequencing. We then sequenced DNA samples from patients with active TB (n = 100) from South Africa on a MinION Mk1C and evaluated the relationship between genotypes and pharmacokinetic parameters for isoniazid (INH) and rifampin (RIF). Measurements and Main Results: The pharmacogenomic panel achieved 100% concordance with Illumina sequencing in variant identification for the samples from the 1,000 Genomes Project. In the clinical cohort, coverage was more than 100× for 1,498 of 1,500 (99.8%) amplicons across the 100 samples. Thirty-three percent, 47%, and 20% of participants were identified as slow, intermediate, and rapid INH acetylators, respectively. INH clearance was 2.2 times higher among intermediate acetylators and 3.8 times higher among rapid acetylators, compared with slow acetylators (P < 0.0001). RIF clearance was 17.3% (2.50-29.9) lower in individuals with homozygous AADAC rs1803155 G→A substitutions (P = 0.0015). Conclusions: Targeted sequencing can enable the detection of polymorphisms that influence TB drug metabolism on a low-cost, portable instrument to personalize dosing for TB treatment or prevention.
Keywords: NAT2; Nanopore; pharmacogenomics; targeted sequencing; tuberculosis.
Figures
Update of
-
A Nanopore sequencing-based pharmacogenomic panel to personalize tuberculosis drug dosing.medRxiv [Preprint]. 2023 Sep 10:2023.09.08.23295248. doi: 10.1101/2023.09.08.23295248. medRxiv. 2023. Update in: Am J Respir Crit Care Med. 2024 Jun 15;209(12):1486-1496. doi: 10.1164/rccm.202309-1583OC. PMID: 37732197 Free PMC article. Updated. Preprint.
Comment in
-
Another Step Closer to Genomic-informed Therapeutic Dosing for Tuberculosis Drugs.Am J Respir Crit Care Med. 2024 Jun 15;209(12):1427-1428. doi: 10.1164/rccm.202403-0566ED. Am J Respir Crit Care Med. 2024. PMID: 38648185 Free PMC article. No abstract available.
References
-
- Merle CS, Fielding K, Sow OB, Gninafon M, Lo MB, Mthiyane T, et al. OFLOTUB/Gatifloxacin for Tuberculosis Project A four-month gatifloxacin-containing regimen for treating tuberculosis. N Engl J Med . 2014;371:1588–1598. - PubMed
-
- Dorman SE, Nahid P, Kurbatova EV, Goldberg SV, Bozeman L, Burman WJ, et al. AIDS Clinical Trials Group and the Tuberculosis Trials Consortium High-dose rifapentine with or without moxifloxacin for shortening treatment of pulmonary tuberculosis: study protocol for TBTC study 31/ACTG A5349 phase 3 clinical trial. Contemp Clin Trials . 2020;90:105938. - PMC - PubMed
-
- Forget EJ, Menzies D. Adverse reactions to first-line antituberculosis drugs. Expert Opin Drug Saf . 2006;5:231–249. - PubMed
-
- Choi H, Park HA, Hyun IG, Kim JH, Hwang YI, Jang SH, et al. Incidence and outcomes of adverse drug reactions to first-line anti-tuberculosis drugs and their effects on the quality of life: a multicenter prospective cohort study. Pharmacoepidemiol Drug Saf . 2022;31:1153–1163. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

