Suppression of Type I Interferon Signaling in Myeloid Cells by Autoantibodies in Severe COVID-19 Patients
- PMID: 38647550
- PMCID: PMC11035476
- DOI: 10.1007/s10875-024-01708-7
Suppression of Type I Interferon Signaling in Myeloid Cells by Autoantibodies in Severe COVID-19 Patients
Abstract
Purpose: Auto-antibodies (auto-abs) to type I interferons (IFNs) have been identified in patients with life-threatening coronavirus disease 2019 (COVID-19), suggesting that the presence of auto-abs may be a risk factor for disease severity. We therefore investigated the mechanism underlying COVID-19 exacerbation induced by auto-abs to type I IFNs.
Methods: We evaluated plasma from 123 patients with COVID-19 to measure auto-abs to type I IFNs. We performed single-cell RNA sequencing (scRNA-seq) of peripheral blood mononuclear cells from the patients with auto-abs and conducted epitope mapping of the auto-abs.
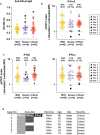
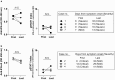
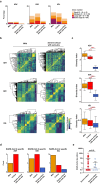
Results: Three of 19 severe and 4 of 42 critical COVID-19 patients had neutralizing auto-abs to type I IFNs. Patients with auto-abs to type I IFNs showed no characteristic clinical features. scRNA-seq from 38 patients with COVID-19 revealed that IFN signaling in conventional dendritic cells and canonical monocytes was attenuated, and SARS-CoV-2-specific BCR repertoires were decreased in patients with auto-abs. Furthermore, auto-abs to IFN-α2 from COVID-19 patients with auto-abs recognized characteristic epitopes of IFN-α2, which binds to the receptor.
Conclusion: Auto-abs to type I IFN found in COVID-19 patients inhibited IFN signaling in dendritic cells and monocytes by blocking the binding of type I IFN to its receptor. The failure to properly induce production of an antibody to SARS-CoV-2 may be a causative factor of COVID-19 severity.
Keywords: Autoantibody; BCR repertoires; COVID-19; Epitope mapping; Single-cell RNA sequencing; Type I IFNs.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Similar Kinetics of Pulmonary SARS-CoV-2 Load in Intensive Care Unit Patients with COVID-19 Pneumonia with or Without Autoantibodies Neutralizing Type I Interferons.J Clin Immunol. 2024 Nov 20;45(1):45. doi: 10.1007/s10875-024-01839-x. J Clin Immunol. 2024. PMID: 39565497
-
Pre-existing Autoantibodies Neutralizing High Concentrations of Type I Interferons in Almost 10% of COVID-19 Patients Admitted to Intensive Care in Barcelona.J Clin Immunol. 2021 Nov;41(8):1733-1744. doi: 10.1007/s10875-021-01136-x. Epub 2021 Sep 27. J Clin Immunol. 2021. PMID: 34570326 Free PMC article.
-
Autoantibodies Neutralizing Type I IFNs in the Bronchoalveolar Lavage of at Least 10% of Patients During Life-Threatening COVID-19 Pneumonia.J Clin Immunol. 2023 Aug;43(6):1093-1103. doi: 10.1007/s10875-023-01512-9. Epub 2023 May 20. J Clin Immunol. 2023. PMID: 37209324 Free PMC article.
-
Type I interferons and SARS-CoV-2: from cells to organisms.Curr Opin Immunol. 2022 Feb;74:172-182. doi: 10.1016/j.coi.2022.01.003. Epub 2022 Jan 25. Curr Opin Immunol. 2022. PMID: 35149239 Free PMC article. Review.
-
Human autoantibodies neutralizing type I IFNs: From 1981 to 2023.Immunol Rev. 2024 Mar;322(1):98-112. doi: 10.1111/imr.13304. Epub 2024 Jan 9. Immunol Rev. 2024. PMID: 38193358 Free PMC article. Review.
Cited by
-
Modulation of antigen delivery and lymph node activation in nonhuman primates by saponin adjuvant saponin/monophosphoryl lipid A nanoparticle.PNAS Nexus. 2024 Nov 25;3(12):pgae529. doi: 10.1093/pnasnexus/pgae529. eCollection 2024 Dec. PNAS Nexus. 2024. PMID: 39677368 Free PMC article.
-
Type I interferon autoantibody footprints reveal neutralizing mechanisms and allow inhibitory decoy design.J Exp Med. 2025 Jun 2;222(6):e20242039. doi: 10.1084/jem.20242039. Epub 2025 Mar 20. J Exp Med. 2025. PMID: 40111224 Free PMC article.
References
-
- Mackenna B, Kennedy NA, Mehrkar A, Rowan A, Galloway J, Matthewman J, et al. Risk of severe COVID-19 outcomes associated with immune-mediated inflammatory diseases and immune-modifying therapies: a nationwide cohort study in the OpenSAFELY platform. The Lancet Rheumatol. 2022;4(7):e490–e506. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- (S) 26221305/Ministry of Education, Culture, Sports, Science and Technology (MEXT Japan) Grants-in-Aid for Scientific Research
- (B) 20H03685/Ministry of Education, Culture, Sports, Science and Technology (MEXT Japan) Grants-in-Aid for Scientific Research
- (C) 17K08876/Ministry of Education, Culture, Sports, Science and Technology (MEXT Japan) Grants-in-Aid for Scientific Research
- (C) 18K07164/Ministry of Education, Culture, Sports, Science and Technology (MEXT Japan) Grants-in-Aid for Scientific Research
- 19K16683/Ministry of Education, Culture, Sports, Science and Technology (MEXT Japan) Grants-in-Aid for Scientific Research
- (B) JP21H05120/Transformative Research Areas
- (B) JP21H05121/Transformative Research Areas
- JP21ek0410060/Practical Research Project for Allergic Diseases and Immunology (Research on Allergic Diseases and Immunology) from the Japan Agency for Medical Research and Development, AMED
- JP21ek0410082/Practical Research Project for Allergic Diseases and Immunology (Research on Allergic Diseases and Immunology) from the Japan Agency for Medical Research and Development, AMED
- JP19ek0410045/Practical Research Project for Allergic Diseases and Immunology (Research on Allergic Diseases and Immunology) from the Japan Agency for Medical Research and Development, AMED
- JP20gm6110005/AMED-PRIME
- JP21gm1210003/AMED-CREST
- JPMJFR200R/JST FOREST Project
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

