Selective biosynthesis of retinol in S. cerevisiae
- PMID: 38647788
- PMCID: PMC10991881
- DOI: 10.1186/s40643-022-00512-8
Selective biosynthesis of retinol in S. cerevisiae
Abstract
The vitamin A component retinol has become an increasingly sought-after cosmetic ingredient. In previous efforts for microbial biosynthesis of vitamin A, a mixture of retinoids was produced. In order to efficiently produce retinol at high purity, the precursor and NADPH supply was first enhanced to improve retinoids accumulation in the S. cerevisiae strain constructed from a β-carotene producer by introducing β-carotene 15,15'-dioxygenase, following by screening of heterologous and endogenous oxidoreductases for retinal reduction. Env9 was found as an endogenous retinal reductase and its activity was verified in vitro. By co-expressing Env9 with the E. coli ybbO, as much as 443.43 mg/L of retinol was produced at 98.76% purity in bi-phasic shake-flask culture when the antioxidant butylated hydroxytoluene was added to prevent retinoids degradation. The retinol titer reached 2479.34 mg/L in fed-batch fermentation. The success in selective biosynthesis of retinol would lay a solid foundation for its biotechnological production.
Keywords: Biosynthesis; Metabolic engineering; Retinal reductase; Retinol; Vitamin A.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
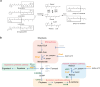
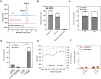
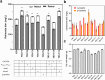
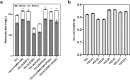
Similar articles
-
Efficient production of retinol in Yarrowia lipolytica by increasing stability using antioxidant and detergent extraction.Metab Eng. 2022 Sep;73:26-37. doi: 10.1016/j.ymben.2022.06.001. Epub 2022 Jun 6. Metab Eng. 2022. PMID: 35671979
-
Metabolic Pathway Coupled with Fermentation Process Optimization for High-Level Production of Retinol in Yarrowia lipolytica.J Agric Food Chem. 2024 Apr 17;72(15):8664-8673. doi: 10.1021/acs.jafc.4c00377. Epub 2024 Apr 2. J Agric Food Chem. 2024. PMID: 38564669
-
Selective production of retinol by engineered Saccharomyces cerevisiae through the expression of retinol dehydrogenase.Biotechnol Bioeng. 2022 Feb;119(2):399-410. doi: 10.1002/bit.28004. Epub 2021 Dec 11. Biotechnol Bioeng. 2022. PMID: 34850377
-
Biochemical properties of retinoid-converting enzymes and biotechnological production of retinoids.Appl Microbiol Biotechnol. 2015 Oct;99(19):7813-26. doi: 10.1007/s00253-015-6830-8. Epub 2015 Aug 1. Appl Microbiol Biotechnol. 2015. PMID: 26231136 Review.
-
Oxidative conversion of carotenoids to retinoids and other products.J Nutr. 2004 Jan;134(1):237S-240S. doi: 10.1093/jn/134.1.237S. J Nutr. 2004. PMID: 14704326 Review.
Cited by
-
Multiplex metabolic engineering of yeast for high-efficiency all-trans-retinoic acid production.Microb Cell Fact. 2025 Jul 4;24(1):157. doi: 10.1186/s12934-025-02782-1. Microb Cell Fact. 2025. PMID: 40615910 Free PMC article.
-
Sustainable biosynthesis of valuable diterpenes in microbes.Eng Microbiol. 2022 Nov 10;3(1):100058. doi: 10.1016/j.engmic.2022.100058. eCollection 2023 Mar. Eng Microbiol. 2022. PMID: 39628524 Free PMC article. Review.
-
Systematic metabolic engineering enables highly efficient production of vitamin A in Saccharomyces cerevisiae.Synth Syst Biotechnol. 2024 Aug 16;10(1):58-67. doi: 10.1016/j.synbio.2024.08.004. eCollection 2025. Synth Syst Biotechnol. 2024. PMID: 39247801 Free PMC article.
-
De novo biosynthesis of nylon 12 monomer ω-aminododecanoic acid.Nat Commun. 2025 Jan 2;16(1):175. doi: 10.1038/s41467-024-55739-0. Nat Commun. 2025. PMID: 39747160 Free PMC article.
-
First report on metagenomics and their predictive functional analysis of fermented bamboo shoot food of Tripura, North East India.Front Microbiol. 2023 Apr 12;14:1158411. doi: 10.3389/fmicb.2023.1158411. eCollection 2023. Front Microbiol. 2023. PMID: 37125168 Free PMC article.
References
-
- Baker ME. Evolution of mammalian 11β- and 17β-hydroxysteroid dehydrogenases-type 2 and retinol dehydrogenases from ancestors in Caenorhabditis elegans and evidence for horizontal transfer of a eukaryote dehydrogenase to E. coli. J Steroid Biochem Mol Biol. 1998;66:355–363. doi: 10.1016/S0960-0760(98)00064-8. - DOI - PubMed
-
- Brachmann CB, Davies A, Cost GJ, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C a useful: set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 2016;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

