Characterization of cold-active trehalose synthase from Pseudarthrobacter sp. for trehalose bioproduction
- PMID: 38647947
- PMCID: PMC10992939
- DOI: 10.1186/s40643-023-00681-0
Characterization of cold-active trehalose synthase from Pseudarthrobacter sp. for trehalose bioproduction
Abstract
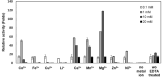
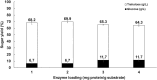
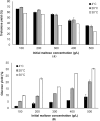
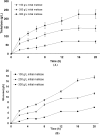
Trehalose is a functional sugar that has numerous applications in food, cosmetic, and pharmaceutical products. Production of trehalose from maltose via a single-step enzymatic catalysis using trehalose synthase (TreS) is a promising method compared with the conventional two-step process due to its simplicity with lower formation of byproducts. In this study, a cold-active trehalose synthase (PaTreS) from Pseudarthrobacter sp. TBRC 2005 was heterologously expressed and characterized. PaTreS showed the maximum activity at 20 °C and maintained 87% and 59% of its activity at 10 °C and 4 °C, respectively. The enzyme had remarkable stability over a board pH range of 7.0-9.0 with the highest activity at pH 7.0. The activity was enhanced by divalent metal ions (Mg2+, Mn2+ and Ca2+). Conversion of high-concentration maltose syrup (100-300 g/L) using PaTreS yielded 71.7-225.5 g/L trehalose, with 4.5-16.4 g/L glucose as a byproduct within 16 h. The work demonstrated the potential of PaTreS as a promising biocatalyst for the development of low-temperature trehalose production, with the advantages of reduced risk of microbial contamination with low generation of byproduct.
Keywords: Pseudarthrobacter sp; Cold active enzyme; Maltose; Trehalose; Trehalose synthase.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Screening, Cloning, Expression and Characterization of New Alkaline Trehalose Synthase from Pseudomonas monteilii and Its Application for Trehalose Production.J Microbiol Biotechnol. 2021 Oct 28;31(10):1455-1464. doi: 10.4014/jmb.2106.06032. J Microbiol Biotechnol. 2021. PMID: 34409951 Free PMC article.
-
Integrated Whole-Cell Biocatalysis for Trehalose Production from Maltose Using Permeabilized Pseudomonas monteilii Cells and Bioremoval of Byproduct.J Microbiol Biotechnol. 2022 Aug 28;32(8):1054-1063. doi: 10.4014/jmb.2202.02028. Epub 2022 May 20. J Microbiol Biotechnol. 2022. PMID: 35791071 Free PMC article.
-
Cloning, expression and functional characterization of a novel trehalose synthase from marine Pseudomonas sp. P8005.World J Microbiol Biotechnol. 2013 Nov;29(11):2195-206. doi: 10.1007/s11274-013-1385-2. Epub 2013 May 29. World J Microbiol Biotechnol. 2013. PMID: 23715900
-
Biotechnical production of trehalose through the trehalose synthase pathway: current status and future prospects.Appl Microbiol Biotechnol. 2018 Apr;102(7):2965-2976. doi: 10.1007/s00253-018-8814-y. Epub 2018 Feb 19. Appl Microbiol Biotechnol. 2018. PMID: 29460000 Review.
-
[Progress on molecular biology of trehalose synthase--a review].Wei Sheng Wu Xue Bao. 2009 Jan;49(1):6-12. Wei Sheng Wu Xue Bao. 2009. PMID: 19388257 Review. Chinese.
Cited by
-
Engineering a high-sugar tolerant strain of Saccharomyces cerevisiae for efficient trehalose production using a cell surface display approach.Bioresour Bioprocess. 2024 Oct 18;11(1):101. doi: 10.1186/s40643-024-00816-x. Bioresour Bioprocess. 2024. PMID: 39422852 Free PMC article.
-
Dietary Trehalose Activates Autophagy Potentially Via MAPK Signaling Pathway to Alleviate Oxidative Stress Induced by Air Exposure Stress in Chinese Mitten Crab (Eriocheir sinensis).Aquac Nutr. 2025 Jul 3;2025:5002677. doi: 10.1155/anu/5002677. eCollection 2025. Aquac Nutr. 2025. PMID: 40642594 Free PMC article.
References
-
- Burek M, Waśkiewicz S, Wandzik I, Kamińska K. Trehalose—properties, biosynthesis and applications. Chemik. 2015;69(8):469–476.
-
- Cai X, Seitl I, Mu W, Zhang T, Stressler T, Fischer L, Jiang B. Characterization of a recombinant trehalose synthase from arthrobacter chlorophenolicus and its unique kinetics indicating a substrate cooperativity. Appl Biochem Biotechnol. 2019;187(4):1255–1271. doi: 10.1007/s12010-018-2877-1. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

