Isolation of xylose-utilizing yeasts from oil palm waste for xylitol and ethanol production
- PMID: 38647966
- PMCID: PMC10992423
- DOI: 10.1186/s40643-023-00691-y
Isolation of xylose-utilizing yeasts from oil palm waste for xylitol and ethanol production
Abstract
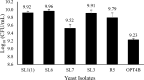
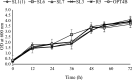
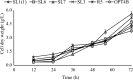
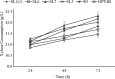
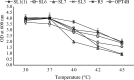
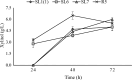
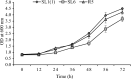
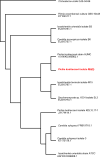
The energy crisis triggers the use of energy sources that are renewable, such as biomass made from lignocellulosic materials, to produce various chemical compounds for food ingredients and biofuel. The efficient conversion of lignocellulosic biomass into products with added value involves the activity of microorganisms, such as yeasts. For the conversion, microorganisms must be able to use various sugars in lignocellulosic biomass, including pentose sugars, especially xylose. This study aims to isolate xylose-utilizing yeasts and analyze their fermentation activity to produce xylitol and ethanol, as well as their ability to grow in liquid hydrolysate produced from pretreated lignocellulosic biomass. Nineteen yeast isolates could grow on solid and liquid media using solely xylose as a carbon source. All isolates can grow in a xylose medium with incubation at 30 °C, 37 °C, 42 °C, and 45 °C. Six isolates, namely SLI (1), SL3, SL6, SL7, R5, and OPT4B, were chosen based on their considerable growth and high xylose consumption rate in a medium with 50 g/L xylose with incubation at 30 °C for 48 h. Four isolates tested, namely SLI (1), SL6, SL7, and R5, can produce xylitol in media containing xylose carbon sources. The concentration of xylitol produced was determined using high-pressure liquid chromatography (HPLC), and the results ranged from 5.0 to 6.0 g/L. Five isolates tested, namely SLI (1), SL6, SL3, R5, and OPT4B, can produce ethanol. The ethanol content produced was determined using gas chromatography (GC), with concentrations ranging from 0.85 to 1.34 g/L. Three isolates, namely SL1(1), R5, and SL6, were able to produce xylitol and ethanol from xylose as carbon sources and were also able to grow on liquid hydrolyzate from pretreated oil palm trunk waste with the subcritical water method. The three isolates were further analyzed using the 18S rDNA sequence to identify the species and confirm their phylogenetic position. Identification based on DNA sequence analysis revealed that isolates SL1(1) and R5 were Pichia kudriavzevii, while isolate SL6 was Candida xylopsoci. The yeast strains isolated from this study could potentially be used for the bioconversion process of lignocellulosic biomass waste to produce value-added derivative products.
Keywords: Ethanol; Lignocellulosic-biomass; Xylitol; Xylose; Yeast.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Cellulolytic enzyme expression and simultaneous conversion of lignocellulosic sugars into ethanol and xylitol by a new Candida tropicalis strain.Biotechnol Biofuels. 2016 Jul 26;9:157. doi: 10.1186/s13068-016-0575-1. eCollection 2016. Biotechnol Biofuels. 2016. PMID: 27462368 Free PMC article.
-
Isolation of xylose-assimilating yeasts and optimization of xylitol production by a new Meyerozyma guilliermondii strain.Int Microbiol. 2020 May;23(2):325-334. doi: 10.1007/s10123-019-00105-0. Epub 2019 Dec 7. Int Microbiol. 2020. PMID: 31813072
-
The production of ethanol from lignocellulosic biomass by Kluyveromyces marxianus CICC 1727-5 and Spathaspora passalidarum ATCC MYA-4345.Appl Microbiol Biotechnol. 2019 Mar;103(6):2845-2855. doi: 10.1007/s00253-019-09625-1. Epub 2019 Jan 31. Appl Microbiol Biotechnol. 2019. PMID: 30706114
-
Genetic improvement of native xylose-fermenting yeasts for ethanol production.J Ind Microbiol Biotechnol. 2015 Jan;42(1):1-20. doi: 10.1007/s10295-014-1535-z. Epub 2014 Nov 18. J Ind Microbiol Biotechnol. 2015. PMID: 25404205 Review.
-
Relation of xylitol formation and lignocellulose degradation in yeast.Appl Microbiol Biotechnol. 2023 May;107(10):3143-3151. doi: 10.1007/s00253-023-12495-3. Epub 2023 Apr 11. Appl Microbiol Biotechnol. 2023. PMID: 37039848 Review.
Cited by
-
Genomic and Functional Analysis of a Novel Yeast Cyberlindnera fabianii TBRC 4498 for High-Yield Xylitol Production.J Fungi (Basel). 2025 Jun 13;11(6):453. doi: 10.3390/jof11060453. J Fungi (Basel). 2025. PMID: 40558965 Free PMC article.
-
Research progress in the biosynthesis of xylitol: feedstock evolution from xylose to glucose.Biotechnol Lett. 2024 Dec;46(6):925-943. doi: 10.1007/s10529-024-03535-7. Epub 2024 Sep 28. Biotechnol Lett. 2024. PMID: 39340754 Review.
References
-
- Almeida JR, Modig T, Petersson A, Hahn-Hägerdal B, Liden G, Gorwa-Grauslund M-F. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol. 2007;82:340–349. doi: 10.1002/JCTB.1676. - DOI
-
- Antunes FAF, Milessi TSS, Chandel AK, Moraes V, Rosa CA, Silva SS. Evaluation of a new yeast from Brazilian biodiversity Scheffersomyces shehatae UFMG-HM 522, for pentose sugars conversion into bioethanol. Biochem Biotechnol Reports. 2013;2(4):1–7. doi: 10.5433/2316-5200.2013v2n4p1. - DOI
-
- Ara´ujo JA, de Abreu-Lima LT, Carreiro SC. Selection and identification of xylose-fermenting yeast strains for ethanol production from lignocellulosic biomass. Boletim Do Centro De Pesquisa De Processamento De Alimentos. 2018;36(1):68–79. doi: 10.5380/bcepplla.v36i1.59557. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

