The genome regulatory landscape of Atlantic salmon liver through smoltification
- PMID: 38648207
- PMCID: PMC11034671
- DOI: 10.1371/journal.pone.0302388
The genome regulatory landscape of Atlantic salmon liver through smoltification
Abstract
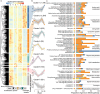
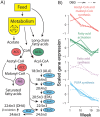
The anadromous Atlantic salmon undergo a preparatory physiological transformation before seawater entry, referred to as smoltification. Key molecular developmental processes involved in this life stage transition, such as remodeling of gill functions, are known to be synchronized and modulated by environmental cues like photoperiod. However, little is known about the photoperiod influence and genome regulatory processes driving other canonical aspects of smoltification such as the large-scale changes in lipid metabolism and energy homeostasis in the developing smolt liver. Here we generate transcriptome, DNA methylation, and chromatin accessibility data from salmon livers across smoltification under different photoperiod regimes. We find a systematic reduction of expression levels of genes with a metabolic function, such as lipid metabolism, and increased expression of energy related genes such as oxidative phosphorylation, during smolt development in freshwater. However, in contrast to similar studies of the gill, smolt liver gene expression prior to seawater transfer was not impacted by photoperiodic history. Integrated analyses of gene expression, chromatin accessibility, and transcription factor (TF) binding signatures highlight chromatin remodeling and TF dynamics underlying smolt gene regulatory changes. Differential peak accessibility patterns largely matched differential gene expression patterns during smoltification and we infer that ZNF682, KLFs, and NFY TFs are important in driving a liver metabolic shift from synthesis to break down of organic compounds in freshwater. Overall, chromatin accessibility and TFBS occupancy were highly correlated to changes in gene expression. On the other hand, we identified numerous differential methylation patterns across the genome, but associated genes were not functionally enriched or correlated to observed gene expression changes across smolt development. Taken together, this work highlights the relative importance of chromatin remodeling during smoltification and demonstrates that metabolic remodeling occurs as a preadaptation to life at sea that is not to a large extent driven by photoperiod history.
Copyright: © 2024 Harvey et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Hoar WS. 4 The Physiology of Smolting Salmonids. In: Hoar WS, Randall DJ, editors. Fish Physiology. 11: Academic Press; 1988. p. 275–343.
-
- McCormick SD, Moriyama S, Björnsson BT. Low temperature limits photoperiod control of smolting in Atlantic salmon through endocrine mechanisms. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology. 2000;278(5):R1352–R61. doi: 10.1152/ajpregu.2000.278.5.R1352 . - DOI - PubMed
-
- Iversen M, Mulugeta T, Gellein Blikeng B, West AC, Jørgensen EH, Rød Sandven S, et al.. RNA profiling identifies novel, photoperiod-history dependent markers associated with enhanced saltwater performance in juvenile Atlantic salmon. PLOS ONE. 2020;15(4):e0227496. doi: 10.1371/journal.pone.0227496 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

