Rhodopsin mislocalization drives ciliary dysregulation in a novel autosomal dominant retinitis pigmentosa knock-in mouse model
- PMID: 38648465
- PMCID: PMC11047207
- DOI: 10.1096/fj.202302260RR
Rhodopsin mislocalization drives ciliary dysregulation in a novel autosomal dominant retinitis pigmentosa knock-in mouse model
Abstract
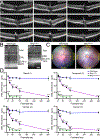
Rhodopsin mislocalization encompasses various blind conditions. Rhodopsin mislocalization is the primary factor leading to rod photoreceptor dysfunction and degeneration in autosomal dominant retinitis pigmentosa (adRP) caused by class I mutations. In this study, we report a new knock-in mouse model that harbors a class I Q344X mutation in the endogenous rhodopsin gene, which causes rod photoreceptor degeneration in an autosomal dominant pattern. In RhoQ344X/+ mice, mRNA transcripts from the wild-type (Rho) and RhoQ344X mutant rhodopsin alleles are expressed at equal levels. However, the amount of RHOQ344X mutant protein is 2.7 times lower than that of wild-type rhodopsin, a finding consistent with the rapid degradation of the mutant protein. Immunofluorescence microscopy indicates that RHOQ344X is mislocalized to the inner segment and outer nuclear layers of rod photoreceptors in both RhoQ344X/+ and RhoQ344X/Q344X mice, confirming the essential role of the C-terminal VxPx motif in promoting OS delivery of rhodopsin. The mislocalization of RHOQ344X is associated with the concurrent mislocalization of wild-type rhodopsin in RhoQ344X/+ mice. To understand the global changes in proteostasis, we conducted quantitative proteomics analysis and found attenuated expression of rod-specific OS membrane proteins accompanying reduced expression of ciliopathy causative gene products, including constituents of BBSome and axonemal dynein subunit. Those studies unveil a novel negative feedback regulation involving ciliopathy-associated proteins. In this process, a defect in the trafficking signal leads to a reduced quantity of the trafficking apparatus, culminating in a widespread reduction in the transport of ciliary proteins.
© 2024 The Authors. The FASEB Journal published by Wiley Periodicals LLC on behalf of Federation of American Societies for Experimental Biology.
Conflict of interest statement
DISCLOSURES
The authors declare no conflicts of interests.
Figures



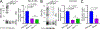
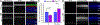

Similar articles
-
P23H rhodopsin accumulation causes transient disruptions to synaptic protein levels in rod photoreceptors in a model of retinitis pigmentosa.Dis Model Mech. 2025 Jun 1;18(6):dmm052256. doi: 10.1242/dmm.052256. Epub 2025 Jun 23. Dis Model Mech. 2025. PMID: 40462724 Free PMC article.
-
Protein kinase G inhibition preserves photoreceptor viability and function in a new mouse model for autosomal dominant retinitis pigmentosa.Cell Death Dis. 2025 Jul 30;16(1):575. doi: 10.1038/s41419-025-07901-9. Cell Death Dis. 2025. PMID: 40738887 Free PMC article.
-
Transcriptional downregulation of rhodopsin is associated with desensitization of rods to light-induced damage in a murine model of retinitis pigmentosa.bioRxiv [Preprint]. 2025 Apr 5:2025.04.03.646684. doi: 10.1101/2025.04.03.646684. bioRxiv. 2025. PMID: 40236225 Free PMC article. Preprint.
-
Prevalence Estimates and Genetic Diversity for Autosomal Dominant Retinitis Pigmentosa Due to RHO, c.68C>A (p.P23H) Variant.Am J Ophthalmol. 2024 Dec;268:340-347. doi: 10.1016/j.ajo.2024.08.038. Epub 2024 Sep 14. Am J Ophthalmol. 2024. PMID: 39278389
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
Cited by
-
Rabin8 phosphorylated by NDR2, the canine early retinal degeneration gene product, directs rhodopsin Golgi-to-cilia trafficking.J Cell Sci. 2025 Jan 15;138(2):JCS263401. doi: 10.1242/jcs.263401. Epub 2025 Jan 23. J Cell Sci. 2025. PMID: 39774853 Free PMC article.
References
-
- Nir I, and Papermaster DS (1986) Immunocytochemical localization of opsin in the inner segment and ciliary plasma membrane of photoreceptors in retinas of rds mutant mice. Invest Ophthalmol Vis Sci 27, 836–840 - PubMed
-
- Bowes C, van Veen T, and Farber DB (1988) Opsin, G-protein and 48-kDa protein in normal and rd mouse retinas: developmental expression of mRNAs and proteins and light/dark cycling of mRNAs. Exp Eye Res 47, 369–390 - PubMed
-
- Nir I, Agarwal N, Sagie G, and Papermaster DS (1989) Opsin distribution and synthesis in degenerating photoreceptors of rd mutant mice. Exp Eye Res 49, 403–421 - PubMed
-
- Lewis GP, Erickson PA, Anderson DH, and Fisher SK (1991) Opsin distribution and protein incorporation in photoreceptors after experimental retinal detachment. Exp Eye Res 53, 629–640 - PubMed
-
- Edward DP, Lim K, Sawaguchi S, and Tso MO (1993) An immunohistochemical study of opsin in photoreceptor cells following light-induced retinal degeneration in the rat. Graefes Arch Clin Exp Ophthalmol 231, 289–294 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

