Porphyrin overdrive rewires cancer cell metabolism
- PMID: 38649187
- PMCID: PMC11035860
- DOI: 10.26508/lsa.202302547
Porphyrin overdrive rewires cancer cell metabolism
Erratum in
-
Correction: Porphyrin overdrive rewires cancer cell metabolism.Life Sci Alliance. 2024 May 20;7(8):e202402816. doi: 10.26508/lsa.202402816. Print 2024 Aug. Life Sci Alliance. 2024. PMID: 38803226 Free PMC article.
Abstract
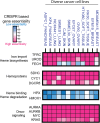
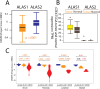
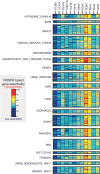
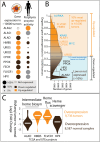
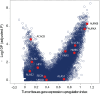
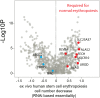
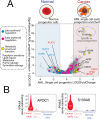
All cancer cells reprogram metabolism to support aberrant growth. Here, we report that cancer cells employ and depend on imbalanced and dynamic heme metabolic pathways, to accumulate heme intermediates, that is, porphyrins. We coined this essential metabolic rewiring "porphyrin overdrive" and determined that it is cancer-essential and cancer-specific. Among the major drivers are genes encoding mid-step enzymes governing the production of heme intermediates. CRISPR/Cas9 editing to engineer leukemia cell lines with impaired heme biosynthetic steps confirmed our whole-genome data analyses that porphyrin overdrive is linked to oncogenic states and cellular differentiation. Although porphyrin overdrive is absent in differentiated cells or somatic stem cells, it is present in patient-derived tumor progenitor cells, demonstrated by single-cell RNAseq, and in early embryogenesis. In conclusion, we identified a dependence of cancer cells on non-homeostatic heme metabolism, and we targeted this cancer metabolic vulnerability with a novel "bait-and-kill" strategy to eradicate malignant cells.
© 2024 Adapa et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







References
-
- Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang C-Z, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, et al. (2016) Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov 6: 914–929. 10.1158/2159-8290.CD-16-0154 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
