Consistent signatures in the human gut microbiome of old- and young-onset colorectal cancer
- PMID: 38649355
- PMCID: PMC11035630
- DOI: 10.1038/s41467-024-47523-x
Consistent signatures in the human gut microbiome of old- and young-onset colorectal cancer
Abstract
The incidence of young-onset colorectal cancer (yCRC) has been increasing in recent decades, but little is known about the gut microbiome of these patients. Most studies have focused on old-onset CRC (oCRC), and it remains unclear whether CRC signatures derived from old patients are valid in young patients. To address this, we assembled the largest yCRC gut metagenomes to date from two independent cohorts and found that the CRC microbiome had limited association with age across adulthood. Differential analysis revealed that well-known CRC-associated taxa, such as Clostridium symbiosum, Peptostreptococcus stomatis, Parvimonas micra and Hungatella hathewayi were significantly enriched (false discovery rate <0.05) in both old- and young-onset patients. Similar strain-level patterns of Fusobacterium nucleatum, Bacteroides fragilis and Escherichia coli were observed for oCRC and yCRC. Almost all oCRC-associated metagenomic pathways had directionally concordant changes in young patients. Importantly, CRC-associated virulence factors (fadA, bft) were enriched in both oCRC and yCRC compared to their respective controls. Moreover, the microbiome-based classification model had similar predication accuracy for CRC status in old- and young-onset patients, underscoring the consistency of microbial signatures across different age groups.
© 2024. The Author(s).
Conflict of interest statement
All authors declare no competing interests.
Figures
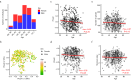



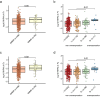

References
-
- Patel SG, Karlitz JJ, Yen T, Lieu CH, Boland CR. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet. Gastroenterol. Hepatol. 2022;7:262–274. doi: 10.1016/S2468-1253(21)00426-X. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

