Tyrosine phosphorylation of CARM1 promotes its enzymatic activity and alters its target specificity
- PMID: 38649367
- PMCID: PMC11035800
- DOI: 10.1038/s41467-024-47689-4
Tyrosine phosphorylation of CARM1 promotes its enzymatic activity and alters its target specificity
Abstract
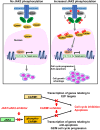
An important epigenetic component of tyrosine kinase signaling is the phosphorylation of histones, and epigenetic readers, writers, and erasers. Phosphorylation of protein arginine methyltransferases (PRMTs), have been shown to enhance and impair their enzymatic activity. In this study, we show that the hyperactivation of Janus kinase 2 (JAK2) by the V617F mutation phosphorylates tyrosine residues (Y149 and Y334) in coactivator-associated arginine methyltransferase 1 (CARM1), an important target in hematologic malignancies, increasing its methyltransferase activity and altering its target specificity. While non-phosphorylatable CARM1 methylates some established substrates (e.g. BAF155 and PABP1), only phospho-CARM1 methylates the RUNX1 transcription factor, on R223 and R319. Furthermore, cells expressing non-phosphorylatable CARM1 have impaired cell-cycle progression and increased apoptosis, compared to cells expressing phosphorylatable, wild-type CARM1, with reduced expression of genes associated with G2/M cell cycle progression and anti-apoptosis. The presence of the JAK2-V617F mutant kinase renders acute myeloid leukemia (AML) cells less sensitive to CARM1 inhibition, and we show that the dual targeting of JAK2 and CARM1 is more effective than monotherapy in AML cells expressing phospho-CARM1. Thus, the phosphorylation of CARM1 by hyperactivated JAK2 regulates its methyltransferase activity, helps select its substrates, and is required for the maximal proliferation of malignant myeloid cells.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

