An increase in ER stress and unfolded protein response in iPSCs-derived neuronal cells from neuronopathic Gaucher disease patients
- PMID: 38649404
- PMCID: PMC11035702
- DOI: 10.1038/s41598-024-59834-6
An increase in ER stress and unfolded protein response in iPSCs-derived neuronal cells from neuronopathic Gaucher disease patients
Abstract
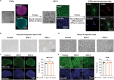
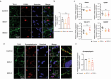
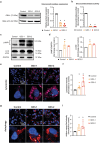
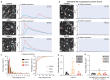
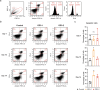
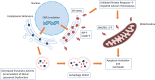
Gaucher disease (GD) is a lysosomal storage disorder caused by a mutation in the GBA1 gene, responsible for encoding the enzyme Glucocerebrosidase (GCase). Although neuronal death and neuroinflammation have been observed in the brains of individuals with neuronopathic Gaucher disease (nGD), the exact mechanism underlying neurodegeneration in nGD remains unclear. In this study, we used two induced pluripotent stem cells (iPSCs)-derived neuronal cell lines acquired from two type-3 GD patients (GD3-1 and GD3-2) to investigate the mechanisms underlying nGD by biochemical analyses. These iPSCs-derived neuronal cells from GD3-1 and GD3-2 exhibit an impairment in endoplasmic reticulum (ER) calcium homeostasis and an increase in unfolded protein response markers (BiP and CHOP), indicating the presence of ER stress in nGD. A significant increase in the BAX/BCL-2 ratio and an increase in Annexin V-positive cells demonstrate a notable increase in apoptotic cell death in GD iPSCs-derived neurons, suggesting downstream signaling after an increase in the unfolded protein response. Our study involves the establishment of iPSCs-derived neuronal models for GD and proposes a possible mechanism underlying nGD. This mechanism involves the activation of ER stress and the unfolded protein response, ultimately leading to apoptotic cell death in neurons.
Keywords: ER stress; Gaucher disease; LSDs; UPR; iPSCs-derived neurons.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Elevated glucosylsphingosine in Gaucher disease induced pluripotent stem cell neurons deregulates lysosomal compartment through mammalian target of rapamycin complex 1.Stem Cells Transl Med. 2021 Jul;10(7):1081-1094. doi: 10.1002/sctm.20-0386. Epub 2021 Mar 3. Stem Cells Transl Med. 2021. PMID: 33656802 Free PMC article.
-
UPR activation and CHOP mediated induction of GBA1 transcription in Gaucher disease.Blood Cells Mol Dis. 2018 Feb;68:21-29. doi: 10.1016/j.bcmd.2016.10.025. Epub 2016 Nov 3. Blood Cells Mol Dis. 2018. PMID: 27856178
-
No evidence for activation of the unfolded protein response in neuronopathic models of Gaucher disease.Hum Mol Genet. 2009 Apr 15;18(8):1482-8. doi: 10.1093/hmg/ddp061. Epub 2009 Feb 3. Hum Mol Genet. 2009. PMID: 19193629
-
Lysosomal functions and dysfunctions: Molecular and cellular mechanisms underlying Gaucher disease and its association with Parkinson disease.Adv Drug Deliv Rev. 2022 Aug;187:114402. doi: 10.1016/j.addr.2022.114402. Epub 2022 Jun 25. Adv Drug Deliv Rev. 2022. PMID: 35764179 Review.
-
Animal Models for the Study of Gaucher Disease.Int J Mol Sci. 2023 Nov 7;24(22):16035. doi: 10.3390/ijms242216035. Int J Mol Sci. 2023. PMID: 38003227 Free PMC article. Review.
Cited by
-
Lysosomal Storage-Independent Fabry Disease Variants with α-Galactosidase A Misprocessing-Induced ER Stress and the Unfolded Protein Response.Nephron. 2025 Mar 20:1-11. doi: 10.1159/000545388. Online ahead of print. Nephron. 2025. PMID: 40112790 Free PMC article. Review.
-
Current opinion on pluripotent stem cell technology in Gaucher's disease: challenges and future prospects.Cytotechnology. 2025 Feb;77(1):26. doi: 10.1007/s10616-024-00687-2. Epub 2024 Dec 27. Cytotechnology. 2025. PMID: 39735330 Review.
-
Establishment of a Human iPSC Line from Mucolipidosis Type II That Expresses the Key Markers of the Disease.Int J Mol Sci. 2025 Apr 19;26(8):3871. doi: 10.3390/ijms26083871. Int J Mol Sci. 2025. PMID: 40332602 Free PMC article.
-
The double whammy of ER-retention and dominant-negative effects in numerous autosomal dominant diseases: significance in disease mechanisms and therapy.J Biomed Sci. 2024 Jun 27;31(1):64. doi: 10.1186/s12929-024-01054-1. J Biomed Sci. 2024. PMID: 38937821 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

