The nematode-trapping fungus Arthrobotrys oligospora detects prey pheromones via G protein-coupled receptors
- PMID: 38649409
- PMCID: PMC11724650
- DOI: 10.1038/s41564-024-01679-w
The nematode-trapping fungus Arthrobotrys oligospora detects prey pheromones via G protein-coupled receptors
Abstract
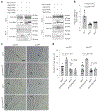
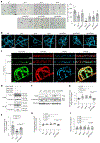
The ability to sense prey-derived cues is essential for predatory lifestyles. Under low-nutrient conditions, Arthrobotrys oligospora and other nematode-trapping fungi develop dedicated structures for nematode capture when exposed to nematode-derived cues, including a conserved family of pheromones, the ascarosides. A. oligospora senses ascarosides via conserved MAPK and cAMP-PKA pathways; however, the upstream receptors remain unknown. Here, using genomic, transcriptomic and functional analyses, we identified two families of G protein-coupled receptors (GPCRs) involved in sensing distinct nematode-derived cues. GPCRs homologous to yeast glucose receptors are required for ascaroside sensing, whereas Pth11-like GPCRs contribute to ascaroside-independent nematode sensing. Both GPCR classes activate conserved cAMP-PKA signalling to trigger trap development. This work demonstrates that predatory fungi use multiple GPCRs to sense several distinct nematode-derived cues for prey recognition and to enable a switch to a predatory lifestyle. Identification of these receptors reveals the molecular mechanisms of cross-kingdom communication via conserved pheromones also sensed by plants and animals.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
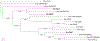



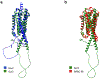
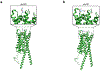




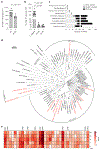


References
-
- Brown NA, Schrevens S, Van Dijck P & Goldman GH Fungal G-protein-coupled receptors: mediators of pathogenesis and targets for disease control. Nat. Microbiol 3, 402–414 (2018). - PubMed
-
- Martín J, Van Den Berg M, van Themaat EVL & Liras P Sensing and transduction of nutritional and chemical signals in filamentous fungi: impact on cell development and secondary metabolites biosynthesis. Biotechnol. Adv 37, 1–15 (2019). - PubMed
-
- Jiang C et al. An expanded subfamily of G-protein-coupled receptor genes in Fusarium graminearum required for wheat infection. Nat. Microbiol 4, 1582–1591 (2019). - PubMed
-
- Johns LE, Goldman GH, Ries LN & Brown NA Nutrient sensing and acquisition in fungi: mechanisms promoting pathogenesis in plant and human hosts. Fungal Biol. Rev 36, 1–14 (2021).
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 110-2311-B-001-047-MY3/Ministry of Science and Technology, Taiwan (Ministry of Science and Technology of Taiwan)
- AS-IA-111-L02/Academia Sinica
- AS-PD 11301-L06/Academia Sinica
- R35GM131877/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R35 GM131877/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources

