Interferon-induced transmembrane protein-1 competitively blocks Ephrin receptor A2-mediated Epstein-Barr virus entry into epithelial cells
- PMID: 38649412
- PMCID: PMC11087256
- DOI: 10.1038/s41564-024-01659-0
Interferon-induced transmembrane protein-1 competitively blocks Ephrin receptor A2-mediated Epstein-Barr virus entry into epithelial cells
Abstract
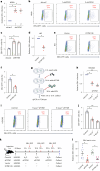
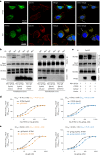
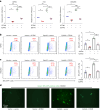
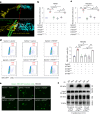
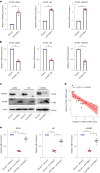
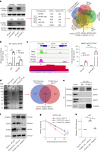
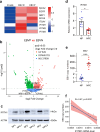
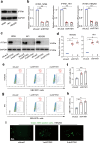
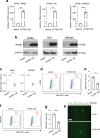
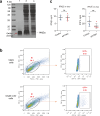
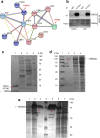
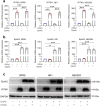
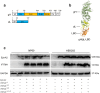
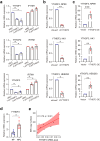
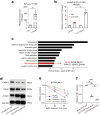
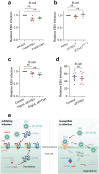
Epstein-Barr virus (EBV) can infect both B cells and epithelial cells (ECs), causing diseases such as mononucleosis and cancer. It enters ECs via Ephrin receptor A2 (EphA2). The function of interferon-induced transmembrane protein-1 (IFITM1) in EBV infection of ECs remains elusive. Here we report that IFITM1 inhibits EphA2-mediated EBV entry into ECs. RNA-sequencing and clinical sample analysis show reduced IFITM1 in EBV-positive ECs and a negative correlation between IFITM1 level and EBV copy number. IFITM1 depletion increases EBV infection and vice versa. Exogenous soluble IFITM1 effectively prevents EBV infection in vitro and in vivo. Furthermore, three-dimensional structure prediction and site-directed mutagenesis demonstrate that IFITM1 interacts with EphA2 via its two specific residues, competitively blocking EphA2 binding to EBV glycoproteins. Finally, YTHDF3, an m6A reader, suppresses IFITM1 via degradation-related DEAD-box protein 5 (DDX5). Thus, this study underscores IFITM1's crucial role in blocking EphA2-mediated EBV entry into ECs, indicating its potential in preventing EBV infection.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

