Mitochondrial enzyme FAHD1 reduces ROS in osteosarcoma
- PMID: 38649439
- PMCID: PMC11035622
- DOI: 10.1038/s41598-024-60012-x
Mitochondrial enzyme FAHD1 reduces ROS in osteosarcoma
Abstract
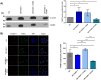
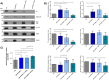
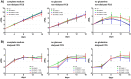
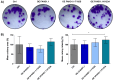
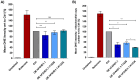
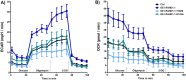
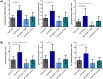
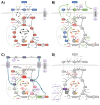
This study investigated the impact of overexpressing the mitochondrial enzyme Fumarylacetoacetate hydrolase domain-containing protein 1 (FAHD1) in human osteosarcoma epithelial cells (U2OS) in vitro. While the downregulation or knockdown of FAHD1 has been extensively researched in various cell types, this study aimed to pioneer the exploration of how increased catalytic activity of human FAHD1 isoform 1 (hFAHD1.1) affects human cell metabolism. Our hypothesis posited that elevation in FAHD1 activity would lead to depletion of mitochondrial oxaloacetate levels. This depletion could potentially result in a decrease in the flux of the tricarboxylic acid (TCA) cycle, thereby accompanied by reduced ROS production. In addition to hFAHD1.1 overexpression, stable U2OS cell lines were established overexpressing a catalytically enhanced variant (T192S) and a loss-of-function variant (K123A) of hFAHD1. It is noteworthy that homologs of the T192S variant are present in animals exhibiting increased resistance to oxidative stress and cancer. Our findings demonstrate that heightened activity of the mitochondrial enzyme FAHD1 decreases cellular ROS levels in U2OS cells. However, these results also prompt a series of intriguing questions regarding the potential role of FAHD1 in mitochondrial metabolism and cellular development.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Identification of human fumarylacetoacetate hydrolase domain-containing protein 1 (FAHD1) as a novel mitochondrial acylpyruvase.J Biol Chem. 2011 Oct 21;286(42):36500-8. doi: 10.1074/jbc.M111.264770. Epub 2011 Aug 30. J Biol Chem. 2011. PMID: 21878618 Free PMC article.
-
Depletion of oxaloacetate decarboxylase FAHD1 inhibits mitochondrial electron transport and induces cellular senescence in human endothelial cells.Exp Gerontol. 2017 Jun;92:7-12. doi: 10.1016/j.exger.2017.03.004. Epub 2017 Mar 8. Exp Gerontol. 2017. PMID: 28286170
-
The mitochondrial enzyme FAHD1 regulates complex II activity in breast cancer cells and is indispensable for basal BT-20 cells in vitro.FEBS Lett. 2022 Nov;596(21):2781-2794. doi: 10.1002/1873-3468.14462. Epub 2022 Aug 12. FEBS Lett. 2022. PMID: 35962472 Free PMC article.
-
Oxaloacetate decarboxylase FAHD1 - a new regulator of mitochondrial function and senescence.Mech Ageing Dev. 2019 Jan;177:22-29. doi: 10.1016/j.mad.2018.07.007. Epub 2018 Jul 25. Mech Ageing Dev. 2019. PMID: 30055189 Review.
-
The Warburg effect in tumor progression: mitochondrial oxidative metabolism as an anti-metastasis mechanism.Cancer Lett. 2015 Jan 28;356(2 Pt A):156-64. doi: 10.1016/j.canlet.2014.04.001. Epub 2014 Apr 13. Cancer Lett. 2015. PMID: 24732809 Free PMC article. Review.
Cited by
-
Single-Cell and Transcriptome Analysis of Periodontitis: Molecular Subtypes and Biomarkers Linked to Mitochondrial Dysfunction and Immunity.J Inflamm Res. 2024 Dec 27;17:11659-11678. doi: 10.2147/JIR.S498739. eCollection 2024. J Inflamm Res. 2024. PMID: 39741754 Free PMC article.
-
FAHD1 and mitochondrial metabolism: a decade of pioneering discoveries.FEBS J. 2025 Jun;292(12):2973-2991. doi: 10.1111/febs.17345. Epub 2024 Dec 6. FEBS J. 2025. PMID: 39642098 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

