Multimodal cell atlas of the ageing human skeletal muscle
- PMID: 38649488
- PMCID: PMC11062927
- DOI: 10.1038/s41586-024-07348-6
Multimodal cell atlas of the ageing human skeletal muscle
Abstract
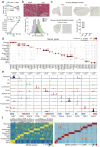
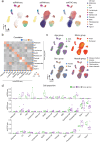
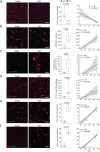
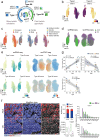
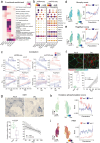
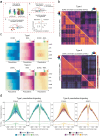
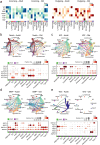
Muscle atrophy and functional decline (sarcopenia) are common manifestations of frailty and are critical contributors to morbidity and mortality in older people1. Deciphering the molecular mechanisms underlying sarcopenia has major implications for understanding human ageing2. Yet, progress has been slow, partly due to the difficulties of characterizing skeletal muscle niche heterogeneity (whereby myofibres are the most abundant) and obtaining well-characterized human samples3,4. Here we generate a single-cell/single-nucleus transcriptomic and chromatin accessibility map of human limb skeletal muscles encompassing over 387,000 cells/nuclei from individuals aged 15 to 99 years with distinct fitness and frailty levels. We describe how cell populations change during ageing, including the emergence of new populations in older people, and the cell-specific and multicellular network features (at the transcriptomic and epigenetic levels) associated with these changes. On the basis of cross-comparison with genetic data, we also identify key elements of chromatin architecture that mark susceptibility to sarcopenia. Our study provides a basis for identifying targets in the skeletal muscle that are amenable to medical, pharmacological and lifestyle interventions in late life.
© 2024. The Author(s).
Conflict of interest statement
Y.L., P.G., J.Z., Y.Y., C.L., L.L., X.X. and M.A.E. are employees of BGI Group. I.R.-P., J.I., E.P., A.L.S. and P.M.-C. are employees of Altos Labs. The other authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

