Single-cell T-cell receptor repertoire profiling in dogs
- PMID: 38649520
- PMCID: PMC11035579
- DOI: 10.1038/s42003-024-06174-w
Single-cell T-cell receptor repertoire profiling in dogs
Abstract
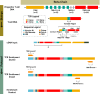
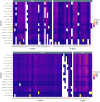
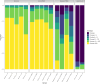
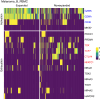
Spontaneous cancers in companion dogs are robust models of human disease. Tracking tumor-specific immune responses in these models requires reagents to perform species-specific single cell T cell receptor sequencing (scTCRseq). scTCRseq and integration with scRNA data have not been demonstrated on companion dogs with cancer. Here, five healthy dogs, two dogs with T cell lymphoma and four dogs with melanoma are selected to demonstrate applicability of scTCRseq in a cancer immunotherapy setting. Single-cell suspensions of PBMCs or lymph node aspirates are profiled using scRNA and dog-specific scTCRseq primers. In total, 77,809 V(D)J-expressing cells are detected, with an average of 3498 (348 - 5,971) unique clonotypes identified per sample. In total, 29/34, 40/40, 22/22 and 9/9 known functional TRAV, TRAJ, TRBV and TRBJ gene segments are observed respectively. Pseudogene or otherwise defective gene segments are also detected supporting re-annotation of several as functional. Healthy dogs exhibit highly diverse repertoires, T cell lymphomas exhibit clonal repertoires, and vaccine-treated melanoma dogs are dominated by a small number of highly abundant clonotypes. scRNA libraries define large clusters of V(D)J-expressing CD8+ and CD4 + T cells. Dominant clonotypes observed in melanoma PBMCs are predominantly CD8 + T cells, with activated phenotypes, suggesting possible anti-tumor T cell populations.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

