Cellular remodeling and JAK inhibition promote zygotic gene expression in the Ciona germline
- PMID: 38649664
- PMCID: PMC11094015
- DOI: 10.1038/s44319-024-00139-0
Cellular remodeling and JAK inhibition promote zygotic gene expression in the Ciona germline
Abstract
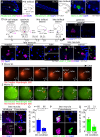
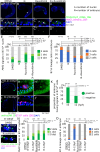
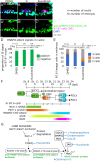
Transcription control is a major determinant of cell fate decisions in somatic tissues. By contrast, early germline fate specification in numerous vertebrate and invertebrate species relies extensively on RNA-level regulation, exerted on asymmetrically inherited maternal supplies, with little-to-no zygotic transcription. However delayed, a maternal-to-zygotic transition is nevertheless poised to complete the deployment of pre-gametic programs in the germline. Here, we focus on early germline specification in the tunicate Ciona to study zygotic genome activation. We first demonstrate that a peculiar cellular remodeling event excludes localized postplasmic Pem-1 mRNA, which encodes the general inhibitor of transcription. Subsequently, zygotic transcription begins in Pem-1-negative primordial germ cells (PGCs), as revealed by histochemical detection of elongating RNA Polymerase II, and nascent Mef2 transcripts. In addition, we uncover a provisional antagonism between JAK and MEK/BMPRI/GSK3 signaling, which controls the onset of zygotic gene expression, following cellular remodeling of PGCs. We propose a 2-step model for the onset of zygotic transcription in the Ciona germline and discuss the significance of germ plasm dislocation and remodeling in the context of developmental fate specification.
Keywords: Germline Specification; Lobe Scission; Primordial Germ Cells; Transcription Control; Tunicate.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Ci-Pem-1 localizes to the nucleus and represses somatic gene transcription in the germline of Ciona intestinalis embryos.Development. 2011 Jul;138(14):2871-81. doi: 10.1242/dev.058131. Development. 2011. PMID: 21693510
-
Control of Pem protein level by localized maternal factors for transcriptional regulation in the germline of the ascidian, Halocynthia roretzi.PLoS One. 2018 Apr 30;13(4):e0196500. doi: 10.1371/journal.pone.0196500. eCollection 2018. PLoS One. 2018. PMID: 29709000 Free PMC article.
-
Microarray analysis of zygotic expression of transcription factor genes and cell signaling molecule genes in early Ciona intestinalis embryos.Dev Growth Differ. 2007 Jan;49(1):27-37. doi: 10.1111/j.1440-169X.2007.00902.x. Dev Growth Differ. 2007. PMID: 17227342
-
The Xenopus Maternal-to-Zygotic Transition from the Perspective of the Germline.Curr Top Dev Biol. 2015;113:271-303. doi: 10.1016/bs.ctdb.2015.07.021. Epub 2015 Aug 21. Curr Top Dev Biol. 2015. PMID: 26358876 Free PMC article. Review.
-
A gene regulatory network for cell fate specification in Ciona embryos.Curr Top Dev Biol. 2020;139:1-33. doi: 10.1016/bs.ctdb.2020.01.001. Epub 2020 May 5. Curr Top Dev Biol. 2020. PMID: 32450958 Review.
Cited by
-
A simple inland culture system provides insights into ascidian post-embryonic developmental physiology.Open Biol. 2025 Jan;15(1):240340. doi: 10.1098/rsob.240340. Epub 2025 Jan 15. Open Biol. 2025. PMID: 39809318 Free PMC article.
-
Cell cycle-driven transcriptome maturation confers multilineage competence to cardiopharyngeal progenitors.bioRxiv [Preprint]. 2024 Jul 23:2024.07.23.604718. doi: 10.1101/2024.07.23.604718. bioRxiv. 2024. PMID: 39091743 Free PMC article. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

