Unveilling genetic profiles and correlations of biofilm-associated genes, quorum sensing, and antibiotic resistance in Staphylococcus aureus isolated from a Malaysian Teaching Hospital
- PMID: 38649897
- PMCID: PMC11036768
- DOI: 10.1186/s40001-024-01831-6
Unveilling genetic profiles and correlations of biofilm-associated genes, quorum sensing, and antibiotic resistance in Staphylococcus aureus isolated from a Malaysian Teaching Hospital
Abstract
Background: Staphylococcus aureus is a notorious multidrug resistant pathogen prevalent in healthcare facilities worldwide. Unveiling the mechanisms underlying biofilm formation, quorum sensing and antibiotic resistance can help in developing more effective therapy for S. aureus infection. There is a scarcity of literature addressing the genetic profiles and correlations of biofilm-associated genes, quorum sensing, and antibiotic resistance among S. aureus isolates from Malaysia.
Methods: Biofilm and slime production of 68 methicillin-susceptible S. aureus (MSSA) and 54 methicillin-resistant (MRSA) isolates were determined using a a plate-based crystal violet assay and Congo Red agar method, respectively. The minimum inhibitory concentration values against 14 antibiotics were determined using VITEK® AST-GP67 cards and interpreted according to CLSI-M100 guidelines. Genetic profiling of 11 S. aureus biofilm-associated genes and agr/sar quorum sensing genes was performed using single or multiplex polymerase chain reaction (PCR) assays.
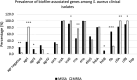
Results: In this study, 75.9% (n = 41) of MRSA and 83.8% (n = 57) of MSSA isolates showed strong biofilm-forming capabilities. Intermediate slime production was detected in approximately 70% of the isolates. Compared to MSSA, significantly higher resistance of clindamycin, erythromycin, and fluoroquinolones was noted among the MRSA isolates. The presence of intracellular adhesion A (icaA) gene was detected in all S. aureus isolates. All MSSA isolates harbored the laminin-binding protein (eno) gene, while all MRSA isolates harbored intracellular adhesion D (icaD), clumping factors A and B (clfA and clfB) genes. The presence of agrI and elastin-binding protein (ebpS) genes was significantly associated with biofilm production in MSSA and MRSA isolates, respectively. In addition, agrI gene was also significantly correlated with oxacillin, cefoxitin, and fluoroquinolone resistance.
Conclusions: The high prevalence of biofilm and slime production among MSSA and MRSA isolates correlates well with the detection of a high prevalence of biofilm-associated genes and agr quorum sensing system. A significant association of agrI gene was found with cefoxitin, oxacillin, and fluoroquinolone resistance. A more focused approach targeting biofilm-associated and quorum sensing genes is important in developing new surveillance and treatment strategies against S. aureus biofilm infection.
Keywords: Antibiotic resistance; Biofilm production; Biofilm-associated gene; MRSA; MSSA.
© 2024. The Author(s).
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
References
-
- Centers of Disease Control and Prevention C. Staphylococcus aureus in healthcare settings 2011. https://www.cdc.gov/hai/organisms/staph.html.
-
- Davis JL. Chapter 2 - Pharmacologic principles. In: Reed SM, Bayly WM, Sellon DC, editors. Equine internal medicine (Fourth Edition) Philadelphia: W.B. Saunders; 2018. pp. 79–137.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous