Dissecting the respective roles of microbiota and host genetics in the susceptibility of Card9-/- mice to colitis
- PMID: 38649950
- PMCID: PMC11036619
- DOI: 10.1186/s40168-024-01798-w
Dissecting the respective roles of microbiota and host genetics in the susceptibility of Card9-/- mice to colitis
Abstract
Background: The etiology of inflammatory bowel disease (IBD) is unclear but involves both genetics and environmental factors, including the gut microbiota. Indeed, exacerbated activation of the gastrointestinal immune system toward the gut microbiota occurs in genetically susceptible hosts and under the influence of the environment. For instance, a majority of IBD susceptibility loci lie within genes involved in immune responses, such as caspase recruitment domain member 9 (Card9). However, the relative impacts of genotype versus microbiota on colitis susceptibility in the context of CARD9 deficiency remain unknown.
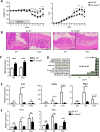
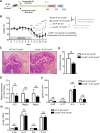
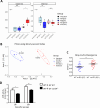
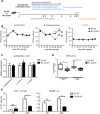
Results: Card9 gene directly contributes to recovery from dextran sodium sulfate (DSS)-induced colitis by inducing the colonic expression of the cytokine IL-22 and the antimicrobial peptides Reg3β and Reg3γ independently of the microbiota. On the other hand, Card9 is required for regulating the microbiota capacity to produce AhR ligands, which leads to the production of IL-22 in the colon, promoting recovery after colitis. In addition, cross-fostering experiments showed that 5 weeks after weaning, the microbiota transmitted from the nursing mother before weaning had a stronger impact on the tryptophan metabolism of the pups than the pups' own genotype.
Conclusions: These results show the role of CARD9 and its effector IL-22 in mediating recovery from DSS-induced colitis in both microbiota-independent and microbiota-dependent manners. Card9 genotype modulates the microbiota metabolic capacity to produce AhR ligands, but this effect can be overridden by the implantation of a WT or "healthy" microbiota before weaning. It highlights the importance of the weaning reaction occurring between the immune system and microbiota for host metabolism and immune functions throughout life. A better understanding of the impact of genetics on microbiota metabolism is key to developing efficient therapeutic strategies for patients suffering from complex inflammatory disorders. Video Abstract.
Keywords: Lactobacillus; AhR ligands; CARD9; Genetics; Gut microbiota; IL-22; Metabolism; Trp metabolism.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Lanternier F, Mahdaviani SA, Barbati E, Chaussade H, Koumar Y, Levy R, Denis B, Brunel A-S, Martin S, Loop M, et al. Inherited CARD9 deficiency in otherwise healthy children and adults with Candida species–induced meningoencephalitis, colitis, or both. Journal of Allergy and Clinical Immunology. 2015;135:1558–1568.e2. doi: 10.1016/j.jaci.2014.12.1930. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

