Transcriptional dynamics and regulatory function of milRNAs in Ascosphaera apis invading Apis mellifera larvae
- PMID: 38650880
- PMCID: PMC11033319
- DOI: 10.3389/fmicb.2024.1355035
Transcriptional dynamics and regulatory function of milRNAs in Ascosphaera apis invading Apis mellifera larvae
Abstract
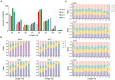
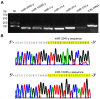
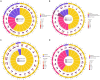
In the present study, small RNA (sRNA) data from Ascosphaera apis were filtered from sRNA-seq datasets from the gut tissues of A. apis-infected Apis mellifera ligustica worker larvae, which were combined with the previously gained sRNA-seq data from A. apis spores to screen differentially expressed milRNAs (DEmilRNAs), followed by trend analysis and investigation of the DEmilRNAs in relation to significant trends. Additionally, the interactions between the DEmilRNAs and their target mRNAs were verified using a dual-luciferase reporter assay. In total, 974 A. apis milRNAs were identified. The first base of these milRNAs was biased toward U. The expression of six milRNAs was confirmed by stem-loop RT-PCR, and the sequences of milR-3245-y and milR-10285-y were validated using Sanger sequencing. These miRNAs grouped into four significant trends, with the target mRNAs of DEmilRNAs involving 42 GO terms and 120 KEGG pathways, such as the fungal-type cell wall and biosynthesis of secondary metabolites. Further investigation demonstrated that 299 DEmilRNAs (novel-m0011-3p, milR-10048-y, bantam-y, etc.) potentially targeted nine genes encoding secondary metabolite-associated enzymes, while 258 (milR-25-y, milR-14-y, milR-932-x, etc.) and 419 (milR-4561-y, milR-10125-y, let-7-x, etc.) DEmilRNAs putatively targeted virulence factor-encoded genes and nine genes involved in the MAPK signaling pathway, respectively. Additionally, the interaction between ADM-B and milR-6882-x, as well as between PKIA and milR-7009-x were verified. Together, these results not only offer a basis for clarifying the mechanisms underlying DEmilRNA-regulated pathogenesis of A. apis and a novel insight into the interaction between A. apis and honey bee larvae, but also provide candidate DEmilRNA-gene axis for further investigation.
Keywords: Apis mellifera; Ascosphaera apis; chalkbrood; infection mechanism; larva; milRNA; regulation.
Copyright © 2024 Fan, Gao, Zang, Liu, Jing, Liu, Guo, Jiang, Wu, Huang, Chen and Guo.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study, the collection, analyses, the interpretation of data, the writing of the manuscript and the decision to publish the results.
Figures





Similar articles
-
Global discovery, expression pattern, and regulatory role of miRNA-like RNAs in Ascosphaera apis infecting the Asian honeybee larvae.Front Microbiol. 2025 Mar 4;16:1551625. doi: 10.3389/fmicb.2025.1551625. eCollection 2025. Front Microbiol. 2025. PMID: 40104596 Free PMC article.
-
Transcriptomic Characterization of miRNAs in Apis cerana Larvae Responding to Ascosphaera apis Infection.Genes (Basel). 2025 Jan 26;16(2):156. doi: 10.3390/genes16020156. Genes (Basel). 2025. PMID: 40004485 Free PMC article.
-
Comprehensive investigation and regulatory function of lncRNAs engaged in western honey bee larval immune response to Ascosphaera apis invasion.Front Physiol. 2022 Dec 16;13:1082522. doi: 10.3389/fphys.2022.1082522. eCollection 2022. Front Physiol. 2022. PMID: 36589426 Free PMC article.
-
First Characterization and Regulatory Function of piRNAs in the Apis mellifera Larval Response to Ascosphaera apis Invasion.Int J Mol Sci. 2023 Nov 15;24(22):16358. doi: 10.3390/ijms242216358. Int J Mol Sci. 2023. PMID: 38003547 Free PMC article.
-
Uncovering the immune responses of Apis mellifera ligustica larval gut to Ascosphaera apis infection utilizing transcriptome sequencing.Gene. 2017 Jul 20;621:40-50. doi: 10.1016/j.gene.2017.04.022. Epub 2017 Apr 18. Gene. 2017. PMID: 28427951
Cited by
-
Global discovery, expression pattern, and regulatory role of miRNA-like RNAs in Ascosphaera apis infecting the Asian honeybee larvae.Front Microbiol. 2025 Mar 4;16:1551625. doi: 10.3389/fmicb.2025.1551625. eCollection 2025. Front Microbiol. 2025. PMID: 40104596 Free PMC article.
References
LinkOut - more resources
Full Text Sources

