The Wolfram-like variant WFS1E864K destabilizes MAM and compromises autophagy and mitophagy in human and mice
- PMID: 38651637
- PMCID: PMC11346566
- DOI: 10.1080/15548627.2024.2341588
The Wolfram-like variant WFS1E864K destabilizes MAM and compromises autophagy and mitophagy in human and mice
Abstract
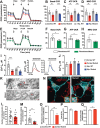
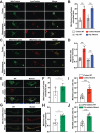
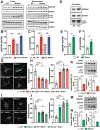
Dominant variants in WFS1 (wolframin ER transmembrane glycoprotein), the gene coding for a mitochondria-associated endoplasmic reticulum (ER) membrane (MAM) resident protein, have been associated with Wolfram-like syndrome (WLS). In vitro and in vivo, WFS1 loss results in reduced ER to mitochondria calcium (Ca2+) transfer, mitochondrial dysfunction, and enhanced macroautophagy/autophagy and mitophagy. However, in the WLS pathological context, whether the mutant protein triggers the same cellular processes is unknown. Here, we show that in human fibroblasts and murine neuronal cultures the WLS protein WFS1E864K leads to decreases in mitochondria bioenergetics and Ca2+ uptake, deregulation of the mitochondrial quality system mechanisms, and alteration of the autophagic flux. Moreover, in the Wfs1E864K mouse, these alterations are concomitant with a decrease of MAM number. These findings reveal pathophysiological similarities between WS and WLS, highlighting the importance of WFS1 for MAM's integrity and functionality. It may open new treatment perspectives for patients with WLS.Abbreviations: BafA1: bafilomycin A1; ER: endoplasmic reticulum; HSPA9/GRP75: heat shock protein family A (Hsp70) member 9; ITPR/IP3R: inositol 1,4,5-trisphosphate receptor; MAM: mitochondria-associated endoplasmic reticulum membrane; MCU: mitochondrial calcium uniporter; MFN2: mitofusin 2; OCR: oxygen consumption rate; ROS: reactive oxygen species; ROT/AA: rotenone+antimycin A; VDAC1: voltage dependent anion channel 1; WLS: Wolfram-like syndrome; WS: Wolfram syndrome; WT: wild-type.
Keywords: Autophagy; WFS1; Wolfram-like syndrome; mitochondria-associated endoplasmic reticulum membrane; mitophagy.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
