The road less taken: Dihydroflavonol 4-reductase inactivation and delphinidin anthocyanin loss underpins a natural intraspecific flower colour variation
- PMID: 38651763
- PMCID: PMC12288823
- DOI: 10.1111/mec.17334
The road less taken: Dihydroflavonol 4-reductase inactivation and delphinidin anthocyanin loss underpins a natural intraspecific flower colour variation
Abstract
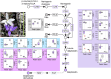
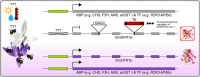
Visual cues are of critical importance for the attraction of animal pollinators, however, little is known about the molecular mechanisms underpinning intraspecific floral colour variation. Here, we combined comparative spectral analysis, targeted metabolite profiling, multi-tissue transcriptomics, differential gene expression, sequence analysis and functional analysis to investigate a bee-pollinated orchid species, Glossodia major with common purple- and infrequent white-flowered morphs. We found uncommon and previously unreported delphinidin-based anthocyanins responsible for the conspicuous and pollinator-perceivable colour of the purple morph and three genetic changes underpinning the loss of colour in the white morph - (1) a loss-of-function (LOF; frameshift) mutation affecting dihydroflavonol 4-reductase (DFR1) coding sequence due to a unique 4-bp insertion, (2) specific downregulation of functional DFR1 expression and (3) the unexpected discovery of chimeric Gypsy transposable element (TE)-gene (DFR) transcripts with potential consequences to the genomic stability and post-transcriptional or epigenetic regulation of DFR. This is one of few known cases where regulatory changes and LOF mutation in an anthocyanin structural gene, rather than transcription factors, are important. Furthermore, if TEs prove to be a frequent source of mutation, the interplay between environmental stress-induced TE evolution and pollinator-mediated selection for adaptive colour variation may be an overlooked mechanism maintaining floral colour polymorphism in nature.
Keywords: anthocyanin; dihydroflavonol 4‐reductase; flower; food deception; orchid; transposable element.
© 2024 The Authors. Molecular Ecology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Afendi, F. M. , Okada, T. , Yamazaki, M. , Hirai‐Morita, A. , Nakamura, Y. , Nakamura, K. , Ikeda, S. , Takahashi, H. , Altaf‐Ul‐Amin, M. , Darusman, L. K. , Saito, K. , & Kanaya, S. (2012). KNApSAcK family databases: Integrated metabolite‐plant species databases for multifaceted plant research. Plant and Cell Physiology, 53, 1–12. - PubMed
-
- Baduel, P. , & Quadrana, L. (2021). Jumpstarting evolution: How transposition can facilitate adaptation to rapid environmental changes. Current Opinion in Plant Biology, 61, 102043. - PubMed
-
- Bontpart, T. , Cheynier, V. , Ageorges, A. , & Terrier, N. (2015). BAHD or SCPL acyltransferase? What a dilemma for acylation in the world of plant phenolic compounds. New Phytologist, 208, 695–707. - PubMed
-
- Borghi, M. , Fernie, A. R. , Schiestl, F. P. , & Bouwmeester, H. J. (2017). The sexual advantage of looking, smelling, and tasting good: The metabolic network that produces signals for pollinators. Trends in Plant Science, 22, 338–350. - PubMed
-
- Buchfink, B. , Xie, C. , & Huson, D. H. (2014). Fast and sensitive protein alignment using DIAMOND. Nature Methods, 12, 59–60. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

