Endogenous tagging using split mNeonGreen in human iPSCs for live imaging studies
- PMID: 38652106
- PMCID: PMC11037917
- DOI: 10.7554/eLife.92819
Endogenous tagging using split mNeonGreen in human iPSCs for live imaging studies
Abstract
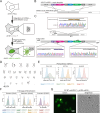
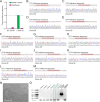
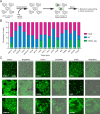
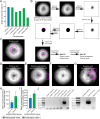
Endogenous tags have become invaluable tools to visualize and study native proteins in live cells. However, generating human cell lines carrying endogenous tags is difficult due to the low efficiency of homology-directed repair. Recently, an engineered split mNeonGreen protein was used to generate a large-scale endogenous tag library in HEK293 cells. Using split mNeonGreen for large-scale endogenous tagging in human iPSCs would open the door to studying protein function in healthy cells and across differentiated cell types. We engineered an iPS cell line to express the large fragment of the split mNeonGreen protein (mNG21-10) and showed that it enables fast and efficient endogenous tagging of proteins with the short fragment (mNG211). We also demonstrate that neural network-based image restoration enables live imaging studies of highly dynamic cellular processes such as cytokinesis in iPSCs. This work represents the first step towards a genome-wide endogenous tag library in human stem cells.
Keywords: cell biology; crispr; cytokinesis; endogenous tagging; gene editing; human; ipsc; live imaging.
Plain language summary
The human body contains around 20,000 different proteins that perform a myriad of essential roles. To understand how these proteins work in healthy individuals and during disease, we need to know their precise locations inside cells and how these locations may change in different situations. Genetic tools known as fluorescent proteins are often used as tags to study the location of specific proteins of interest within cells. When exposed to light, the fluorescent proteins emit specific colours of light that can be observed using microscopes. In a fluorescent protein system known as split mNeonGreen, researchers insert the DNA encoding two fragments of a fluorescent protein (one large, one small) separately into cells. The large fragment can be found throughout the cell, while the small fragment is attached to specific host proteins. When the two fragments meet, they assemble into the full mNeonGreen protein and can fluoresce. Researchers can use split mNeonGreen and other similar systems to generate large libraries of cells where the small fragment of a fluorescent protein is attached to thousands of different host proteins. However, so far these libraries are restricted to a handful of different types of cells. To address this challenge, Husser et al. inserted the DNA encoding the large fragment of mNeonGreen into human cells known as induced pluripotent stem cells, which are able to give rise to any other type of human cell. This then enabled the team to quickly and efficiently generate a library of stem cells that express the small fragment of mNeonGreen attached to different host proteins. Further experiments studied the locations of host proteins in the stem cells just before they divided into two cells. This suggested that there are differences between how induced pluripotent stem cells and other types of cells divide. In the future, the cells and the method developed by Husser et al. may be used by other researchers to create atlases showing where human proteins are located in many other types of cells.
© 2023, Husser et al.
Conflict of interest statement
MH, NP, CL, FA, VM, AP No competing interests declared
Figures





Update of
- doi: 10.1101/2023.06.28.546942
- doi: 10.7554/eLife.92819.1
- doi: 10.7554/eLife.92819.2
Similar articles
-
Improved yellow-green split fluorescent proteins for protein labeling and signal amplification.PLoS One. 2020 Nov 23;15(11):e0242592. doi: 10.1371/journal.pone.0242592. eCollection 2020. PLoS One. 2020. PMID: 33227014 Free PMC article.
-
Tissue-specific and endogenous protein labeling with split fluorescent proteins.bioRxiv [Preprint]. 2024 Feb 28:2024.02.28.581822. doi: 10.1101/2024.02.28.581822. bioRxiv. 2024. Update in: Dev Biol. 2024 Oct;514:109-116. doi: 10.1016/j.ydbio.2024.06.011. PMID: 38464062 Free PMC article. Updated. Preprint.
-
Endogenous Protein Tagging in Human Induced Pluripotent Stem Cells Using CRISPR/Cas9.J Vis Exp. 2018 Aug 25;(138):58130. doi: 10.3791/58130. J Vis Exp. 2018. PMID: 30199041 Free PMC article.
-
Gene correction in patient-specific iPSCs for therapy development and disease modeling.Hum Genet. 2016 Sep;135(9):1041-58. doi: 10.1007/s00439-016-1691-5. Epub 2016 Jun 2. Hum Genet. 2016. PMID: 27256364 Free PMC article. Review.
-
Genome Editing in Induced Pluripotent Stem Cells using CRISPR/Cas9.Stem Cell Rev Rep. 2018 Jun;14(3):323-336. doi: 10.1007/s12015-018-9811-3. Stem Cell Rev Rep. 2018. PMID: 29623532 Review.
Cited by
-
Efficient endogenous protein labelling in Dictyostelium using CRISPR/Cas9 knock-in and split fluorescent proteins.PLoS One. 2025 Jun 20;20(6):e0326577. doi: 10.1371/journal.pone.0326577. eCollection 2025. PLoS One. 2025. PMID: 40540504 Free PMC article.
References
-
- Araujo F. Nanopore Demultiplex with Bowtie. c85a2ccGitHub. 2022 https://github.com/frba/nanopore_demultiplex
-
- Bhargava N, Thakur P, Muruganandam TP, Jaitly S, Gupta P, Lohani N, Goswami SG, Saravanakumar V, Bhattacharya SK, Jain S, Ramalingam S. Development of an efficient single-cell cloning and expansion strategy for genome edited induced pluripotent stem cells. Molecular Biology Reports. 2022;49:7887–7898. doi: 10.1007/s11033-022-07621-9. - DOI - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
- Doctoral Training Scholarship B2X/Fonds de recherche du Québec - Nature et technologies
- SynBioApps scholarship/Concordia University
- Disruptive Technology Solutions for Cell and Gene Therapy Challenge/National Research Council Canada
- Discovery Grant/Natural Sciences and Engineering Research Council of Canada
- RGPIN-2023-04805/Natural Sciences and Engineering Research Council of Canada
LinkOut - more resources
Full Text Sources
Research Materials

