Human NLRC4 expression promotes cancer survival and associates with type I interferon signaling and immune infiltration
- PMID: 38652550
- PMCID: PMC11142746
- DOI: 10.1172/JCI166085
Human NLRC4 expression promotes cancer survival and associates with type I interferon signaling and immune infiltration
Abstract
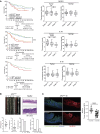
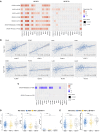
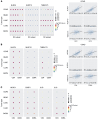
The immune system can control cancer progression. However, even though some innate immune sensors of cellular stress are expressed intrinsically in epithelial cells, their potential role in cancer aggressiveness and subsequent overall survival in humans is mainly unknown. Here, we show that nucleotide-binding oligomerization domain-like receptor (NLR) family CARD domain-containing 4 (NLRC4) is downregulated in epithelial tumor cells of patients with colorectal cancer (CRC) by using spatial tissue imaging. Strikingly, only the loss of tumor NLRC4, but not stromal NLRC4, was associated with poor immune infiltration (mainly DCs and CD4+ and CD8+ T cells) and accurately predicted progression to metastatic stage IV and decrease in overall survival. By combining multiomics approaches, we show that restoring NLRC4 expression in human CRC cells triggered a broad inflammasome-independent immune reprogramming consisting of type I interferon (IFN) signaling genes and the release of chemokines and myeloid growth factors involved in the tumor infiltration and activation of DCs and T cells. Consistently, such reprogramming in cancer cells was sufficient to directly induce maturation of human DCs toward a Th1 antitumor immune response through IL-12 production in vitro. In multiple human carcinomas (colorectal, lung, and skin), we confirmed that NLRC4 expression in patient tumors was strongly associated with type I IFN genes, immune infiltrates, and high microsatellite instability. Thus, we shed light on the epithelial innate immune sensor NLRC4 as a therapeutic target to promote an efficient antitumor immune response against the aggressiveness of various carcinomas.
Keywords: Cancer immunotherapy; Cellular immune response; Immunology; Innate immunity; Oncology.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

