HSV-1 employs UL56 to antagonize expression and function of cGAMP channels
- PMID: 38652659
- PMCID: PMC7617956
- DOI: 10.1016/j.celrep.2024.114122
HSV-1 employs UL56 to antagonize expression and function of cGAMP channels
Abstract
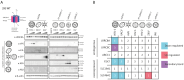
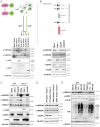
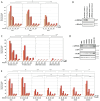
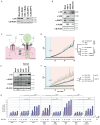
DNA sensing is important for antiviral immunity. The DNA sensor cGAS synthesizes 2'3'-cyclic GMP-AMP (cGAMP), a second messenger that activates STING, which induces innate immunity. cGAMP not only activates STING in the cell where it is produced but cGAMP also transfers to other cells. Transporters, channels, and pores (including SLC19A1, SLC46A2, P2X7, ABCC1, and volume-regulated anion channels (VRACs)) release cGAMP into the extracellular space and/or import cGAMP. We report that infection with multiple human viruses depletes some of these cGAMP conduits. This includes herpes simplex virus 1 (HSV-1) that targets SLC46A2, P2X7, and the VRAC subunits LRRC8A and LRRC8C for degradation. The HSV-1 protein UL56 is necessary and sufficient for these effects that are mediated at least partially by proteasomal turnover. UL56 thereby inhibits cGAMP uptake via VRAC, SLC46A2, and P2X7. Taken together, HSV-1 antagonizes intercellular cGAMP transfer. We propose that this limits innate immunity by reducing cell-to-cell communication via the immunotransmitter cGAMP.
Keywords: CP: Immunology; CP: Microbiology; DNA sensing; LRRC8A; P2X7; SLC46A2; STING; UL56; VRAC; cGAMP; cGAMP transport; herpes simplex virus.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Medzhitov R. Approaching the asymptote: 20 years later. Immunity. 2009;30:766–775. - PubMed
-
- Ablasser A, Chen ZJ. cGAS in action: Expanding roles in immunity and inflammation. Science. 2019;363:eaat8657. - PubMed
-
- Guey B, Ablasser A. Emerging dimensions of cellular cGAS-STING signaling. Curr Opin Immunol. 2022;74:164–171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

