Conserved Genes in Highly Regenerative Metazoans Are Associated with Planarian Regeneration
- PMID: 38652806
- PMCID: PMC11077316
- DOI: 10.1093/gbe/evae082
Conserved Genes in Highly Regenerative Metazoans Are Associated with Planarian Regeneration
Abstract
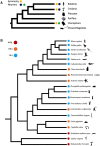
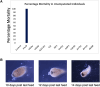
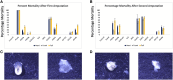
Metazoan species depict a wide spectrum of regeneration ability which calls into question the evolutionary origins of the underlying processes. Since species with high regeneration ability are widely distributed throughout metazoans, there is a possibility that the metazoan ancestor had an underlying common molecular mechanism. Early metazoans like sponges possess high regenerative ability, but, due to the large differences they have with Cnidaria and Bilateria regarding symmetry and neuronal systems, it can be inferred that this regenerative ability is different. We hypothesized that the last common ancestor of Cnidaria and Bilateria possessed remarkable regenerative ability which was lost during evolution. We separated Cnidaria and Bilateria into three classes possessing whole-body regenerating, high regenerative ability, and low regenerative ability. Using a multiway BLAST and gene phylogeny approach, we identified genes conserved in whole-body regenerating species and lost in low regenerative ability species and labeled them Cnidaria and Bilaterian regeneration genes. Through transcription factor analysis, we identified that Cnidaria and Bilaterian regeneration genes were associated with an overabundance of homeodomain regulatory elements. RNA interference of Cnidaria and Bilaterian regeneration genes resulted in loss of regeneration phenotype for HRJDa, HRJDb, DUF21, DISP3, and ARMR genes. We observed that DUF21 knockdown was highly lethal in the early stages of regeneration indicating a potential role in wound response. Also, HRJDa, HRJDb, DISP3, and ARMR knockdown showed loss of regeneration phenotype after second amputation. The results strongly correlate with their respective RNA-seq profiles. We propose that Cnidaria and Bilaterian regeneration genes play a major role in regeneration across highly regenerative Cnidaria and Bilateria.
Keywords: comparative genomics; planarian; regeneration.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

Similar articles
-
JmjC Domain-Encoding Genes Are Conserved in Highly Regenerative Metazoans and Are Associated with Planarian Whole-Body Regeneration.Genome Biol Evol. 2019 Feb 1;11(2):552-564. doi: 10.1093/gbe/evz021. Genome Biol Evol. 2019. PMID: 30698705 Free PMC article.
-
Initiating a regenerative response; cellular and molecular features of wound healing in the cnidarian Nematostella vectensis.BMC Biol. 2014 Mar 26;12:24. doi: 10.1186/1741-7007-12-24. BMC Biol. 2014. PMID: 24670243 Free PMC article.
-
Colonial origin for Emetazoa: major morphological transitions and the origin of bilaterian complexity.J Morphol. 2000 Jan;243(1):35-74. doi: 10.1002/(SICI)1097-4687(200001)243:1<35::AID-JMOR3>3.0.CO;2-#. J Morphol. 2000. PMID: 10629096 Review.
-
Ancient origins of axial patterning genes: Hox genes and ParaHox genes in the Cnidaria.Evol Dev. 1999 Jul-Aug;1(1):16-23. doi: 10.1046/j.1525-142x.1999.99010.x. Evol Dev. 1999. PMID: 11324016
-
The use of planarians to dissect the molecular basis of metazoan regeneration.Wound Repair Regen. 1998 Jul-Aug;6(4):413-20. doi: 10.1046/j.1524-475x.1998.60418.x. Wound Repair Regen. 1998. PMID: 9824561 Review.
Cited by
-
From fragment to form: whole-body regeneration in a model urochordate.NPJ Regen Med. 2025 Aug 1;10(1):36. doi: 10.1038/s41536-025-00423-0. NPJ Regen Med. 2025. PMID: 40750611 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

