Gene-specific somatic epigenetic mosaicism of FDFT1 underlies a non-hereditary localized form of porokeratosis
- PMID: 38653249
- PMCID: PMC11080608
- DOI: 10.1016/j.ajhg.2024.03.017
Gene-specific somatic epigenetic mosaicism of FDFT1 underlies a non-hereditary localized form of porokeratosis
Abstract
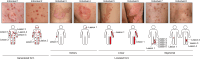
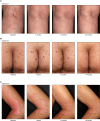
Porokeratosis is a clonal keratinization disorder characterized by solitary, linearly arranged, or generally distributed multiple skin lesions. Previous studies showed that genetic alterations in MVK, PMVK, MVD, or FDPS-genes in the mevalonate pathway-cause hereditary porokeratosis, with skin lesions harboring germline and lesion-specific somatic variants on opposite alleles. Here, we identified non-hereditary porokeratosis associated with epigenetic silencing of FDFT1, another gene in the mevalonate pathway. Skin lesions of the generalized form had germline and lesion-specific somatic variants on opposite alleles in FDFT1, representing FDFT1-associated hereditary porokeratosis identified in this study. Conversely, lesions of the solitary or linearly arranged localized form had somatic bi-allelic promoter hypermethylation or mono-allelic promoter hypermethylation with somatic genetic alterations on opposite alleles in FDFT1, indicating non-hereditary porokeratosis. FDFT1 localization was uniformly diminished within the lesions, and lesion-derived keratinocytes showed cholesterol dependence for cell growth and altered expression of genes related to cell-cycle and epidermal development, confirming that lesions form by clonal expansion of FDFT1-deficient keratinocytes. In some individuals with the localized form, gene-specific promoter hypermethylation of FDFT1 was detected in morphologically normal epidermis adjacent to methylation-related lesions but not distal to these lesions, suggesting that asymptomatic somatic epigenetic mosaicism of FDFT1 predisposes certain skin areas to the disease. Finally, consistent with its genetic etiology, topical statin treatment ameliorated lesions in FDFT1-deficient porokeratosis. In conclusion, we identified bi-allelic genetic and/or epigenetic alterations of FDFT1 as a cause of porokeratosis and shed light on the pathogenesis of skin mosaicism involving clonal expansion of epigenetically altered cells.
Keywords: FDFT1; cholesterol; clonal expansion; epigenetic mosaicism; germline variant; mevalonate pathway; porokeratosis; promoter hypermethylation; somatic variant; statin.
Copyright © 2024 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests T.T. and M.I. are employees of Japan Tissue Engineering Co., Ltd.
Figures






Similar articles
-
Clonal Expansion of Second-Hit Cells with Somatic Recombinations or C>T Transitions Form Porokeratosis in MVD or MVK Mutant Heterozygotes.J Invest Dermatol. 2019 Dec;139(12):2458-2466.e9. doi: 10.1016/j.jid.2019.05.020. Epub 2019 Jun 15. J Invest Dermatol. 2019. PMID: 31207227
-
Second-Hit, Postzygotic PMVK and MVD Mutations in Linear Porokeratosis.JAMA Dermatol. 2019 May 1;155(5):548-555. doi: 10.1001/jamadermatol.2019.0016. JAMA Dermatol. 2019. PMID: 30942823 Free PMC article.
-
Mutations in mevalonate pathway genes in patients with familial or sporadic porokeratosis.J Dermatol. 2018 Jul;45(7):862-866. doi: 10.1111/1346-8138.14343. Epub 2018 May 3. J Dermatol. 2018. PMID: 29722423
-
Twists and turns of the genetic story of mevalonate kinase-associated diseases: A review.Genes Dis. 2021 Jun 9;9(4):1000-1007. doi: 10.1016/j.gendis.2021.05.002. eCollection 2022 Jul. Genes Dis. 2021. PMID: 35685471 Free PMC article. Review.
-
Cancer proneness of linear porokeratosis may be explained by allelic loss.Dermatology. 1997;195(1):20-5. doi: 10.1159/000245678. Dermatology. 1997. PMID: 9267731 Review.
Cited by
-
Somatic mutations in arteriovenous malformations in hereditary hemorrhagic telangiectasia support a bi-allelic two-hit mutation mechanism of pathogenesis.Am J Hum Genet. 2024 Oct 3;111(10):2283-2298. doi: 10.1016/j.ajhg.2024.08.020. Epub 2024 Sep 18. Am J Hum Genet. 2024. PMID: 39299239 Free PMC article.
-
The Role of Somatic Mutation in Hereditary Hemorrhagic Telangiectasia Pathogenesis.J Clin Med. 2025 Jun 24;14(13):4479. doi: 10.3390/jcm14134479. J Clin Med. 2025. PMID: 40648852 Free PMC article. Review.
-
Advanced phasing techniques in congenital skin diseases.J Dermatol. 2025 Mar;52(3):392-399. doi: 10.1111/1346-8138.17597. Epub 2024 Dec 26. J Dermatol. 2025. PMID: 39723554 Free PMC article. Review.
-
Improved multiancestry fine-mapping identifies cis-regulatory variants underlying molecular traits and disease risk.Nat Genet. 2025 Aug;57(8):1881-1889. doi: 10.1038/s41588-025-02262-7. Epub 2025 Jul 21. Nat Genet. 2025. PMID: 40691406
-
Considerations for reporting variants in novel candidate genes identified during clinical genomic testing.bioRxiv [Preprint]. 2024 Jun 21:2024.02.05.579012. doi: 10.1101/2024.02.05.579012. bioRxiv. 2024. Update in: Genet Med. 2024 Oct;26(10):101199. doi: 10.1016/j.gim.2024.101199. PMID: 38370830 Free PMC article. Updated. Preprint.
References
-
- Biesecker L.G., Spinner N.B. A genomic view of mosaicism and human disease. Nat. Rev. Genet. 2013;14:307–320. - PubMed
-
- Happle R. Mosaicism in human skin. Understanding the patterns and mechanisms. Arch. Dermatol. 1993;129:1460–1470. - PubMed
-
- Reed R.J., Leone P. Porokeratosis--a mutant clonal keratosis of the epidermis. I. Histogenesis. Arch. Dermatol. 1970;101:340–347. - PubMed
-
- Das A., Vasudevan B., Talwar A. Porokeratosis: An enigma beginning to unravel. Indian J. Dermatol. Venereol. Leprol. 2022;88:291–299. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

