Targeting NRAS via miR-1304-5p or farnesyltransferase inhibition confers sensitivity to ALK inhibitors in ALK-mutant neuroblastoma
- PMID: 38653965
- PMCID: PMC11039739
- DOI: 10.1038/s41467-024-47771-x
Targeting NRAS via miR-1304-5p or farnesyltransferase inhibition confers sensitivity to ALK inhibitors in ALK-mutant neuroblastoma
Abstract
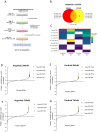
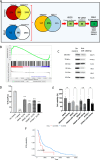
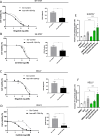
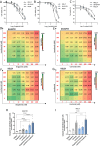
Targeting Anaplastic lymphoma kinase (ALK) is a promising therapeutic strategy for aberrant ALK-expressing malignancies including neuroblastoma, but resistance to ALK tyrosine kinase inhibitors (ALK TKI) is a distinct possibility necessitating drug combination therapeutic approaches. Using high-throughput, genome-wide CRISPR-Cas9 knockout screens, we identify miR-1304-5p loss as a desensitizer to ALK TKIs in aberrant ALK-expressing neuroblastoma; inhibition of miR-1304-5p decreases, while mimics of this miRNA increase the sensitivity of neuroblastoma cells to ALK TKIs. We show that miR-1304-5p targets NRAS, decreasing cell viability via induction of apoptosis. It follows that the farnesyltransferase inhibitor (FTI) lonafarnib in addition to ALK TKIs act synergistically in neuroblastoma, inducing apoptosis in vitro. In particular, on combined treatment of neuroblastoma patient derived xenografts with an FTI and an ALK TKI complete regression of tumour growth is observed although tumours rapidly regrow on cessation of therapy. Overall, our data suggests that combined use of ALK TKIs and FTIs, constitutes a therapeutic approach to treat high risk neuroblastoma although prolonged therapy is likely required to prevent relapse.
© 2024. The Author(s).
Conflict of interest statement
G.A.A.B. has received institutional consultancy fees from Roche, Takeda, Novartis, and Janssen. All other authors have no conflicts of interest to declare.
Figures

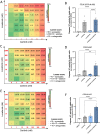
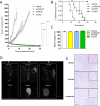
References
-
- Friboulet L, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014;4:662–673. doi: 10.1158/2159-8290.CD-13-0846. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

