Temporal coordination of the transcription factor response to H2O2 stress
- PMID: 38653977
- PMCID: PMC11039679
- DOI: 10.1038/s41467-024-47837-w
Temporal coordination of the transcription factor response to H2O2 stress
Abstract
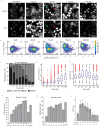
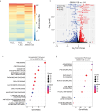
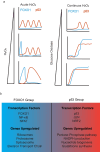
Oxidative stress from excess H2O2 activates transcription factors that restore redox balance and repair oxidative damage. Although many transcription factors are activated by H2O2, it is unclear whether they are activated at the same H2O2 concentration, or time. Dose-dependent activation is likely as oxidative stress is not a singular state and exhibits dose-dependent outcomes including cell-cycle arrest and cell death. Here, we show that transcription factor activation is both dose-dependent and coordinated over time. Low levels of H2O2 activate p53, NRF2 and JUN. Yet under high H2O2, these transcription factors are repressed, and FOXO1, NF-κB, and NFAT1 are activated. Time-lapse imaging revealed that the order in which these two groups of transcription factors are activated depends on whether H2O2 is administered acutely by bolus addition, or continuously through the glucose oxidase enzyme. Finally, we provide evidence that 2-Cys peroxiredoxins control which group of transcription factors are activated.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Update of
-
Temporal Coordination of the Transcription Factor Response to H2O2 stress.bioRxiv [Preprint]. 2023 Mar 7:2023.03.07.531593. doi: 10.1101/2023.03.07.531593. bioRxiv. 2023. Update in: Nat Commun. 2024 Apr 23;15(1):3440. doi: 10.1038/s41467-024-47837-w. PMID: 36945409 Free PMC article. Updated. Preprint.
-
Temporal Coordination of the Transcription Factor Response to H2O2 stress.Res Sq [Preprint]. 2023 May 4:rs.3.rs-2791121. doi: 10.21203/rs.3.rs-2791121/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Apr 23;15(1):3440. doi: 10.1038/s41467-024-47837-w. PMID: 37205449 Free PMC article. Updated. Preprint.
Similar articles
-
Temporal Coordination of the Transcription Factor Response to H2O2 stress.Res Sq [Preprint]. 2023 May 4:rs.3.rs-2791121. doi: 10.21203/rs.3.rs-2791121/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Apr 23;15(1):3440. doi: 10.1038/s41467-024-47837-w. PMID: 37205449 Free PMC article. Updated. Preprint.
-
Temporal Coordination of the Transcription Factor Response to H2O2 stress.bioRxiv [Preprint]. 2023 Mar 7:2023.03.07.531593. doi: 10.1101/2023.03.07.531593. bioRxiv. 2023. Update in: Nat Commun. 2024 Apr 23;15(1):3440. doi: 10.1038/s41467-024-47837-w. PMID: 36945409 Free PMC article. Updated. Preprint.
-
Mechanisms underlying Nrf2 nuclear translocation by non-lethal levels of hydrogen peroxide: p38 MAPK-dependent neutral sphingomyelinase2 membrane trafficking and ceramide/PKCζ/CK2 signaling.Free Radic Biol Med. 2022 Oct;191:191-202. doi: 10.1016/j.freeradbiomed.2022.08.036. Epub 2022 Sep 3. Free Radic Biol Med. 2022. PMID: 36064071 Review.
-
The Mitochondrial-Derived Peptide MOTS-c Attenuates Oxidative Stress Injury and the Inflammatory Response of H9c2 Cells Through the Nrf2/ARE and NF-κB Pathways.Cardiovasc Eng Technol. 2022 Oct;13(5):651-661. doi: 10.1007/s13239-021-00589-w. Epub 2021 Dec 2. Cardiovasc Eng Technol. 2022. PMID: 34859377
-
The effects of chromium(VI) on the thioredoxin system: implications for redox regulation.Free Radic Biol Med. 2012 May 15;52(10):2091-107. doi: 10.1016/j.freeradbiomed.2012.03.013. Epub 2012 Apr 18. Free Radic Biol Med. 2012. PMID: 22542445 Free PMC article. Review.
Cited by
-
H2O2 accumulation promoting internalization of ox-LDL in early atherosclerosis revealed via a synergistic dual-functional NIR fluorescence probe.Chem Sci. 2024 Nov 23;16(1):345-353. doi: 10.1039/d4sc05546b. eCollection 2024 Dec 18. Chem Sci. 2024. PMID: 39620070 Free PMC article.
-
Fundamentals of redox regulation in biology.Nat Rev Mol Cell Biol. 2024 Sep;25(9):701-719. doi: 10.1038/s41580-024-00730-2. Epub 2024 Apr 30. Nat Rev Mol Cell Biol. 2024. PMID: 38689066 Free PMC article. Review.
-
Harnessing the interaction between redox signaling and senescence to restrain tumor drug resistance.Front Cell Dev Biol. 2025 Jul 9;13:1639772. doi: 10.3389/fcell.2025.1639772. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40703657 Free PMC article. Review.
-
Oxidative stress controls lncRNA-mediated sow granulosa cell functions in a FoxO1-dependent manner.J Anim Sci Biotechnol. 2024 Dec 16;15(1):171. doi: 10.1186/s40104-024-01120-6. J Anim Sci Biotechnol. 2024. PMID: 39681884 Free PMC article.
-
MLIP and Its Potential Influence on Key Oncogenic Pathways.Cells. 2024 Jun 26;13(13):1109. doi: 10.3390/cells13131109. Cells. 2024. PMID: 38994962 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

