Genome-wide identification of ZmMYC2 binding sites and target genes in maize
- PMID: 38654166
- PMCID: PMC11036654
- DOI: 10.1186/s12864-024-10297-z
Genome-wide identification of ZmMYC2 binding sites and target genes in maize
Erratum in
-
Correction: Genome-wide identification of ZmMYC2 binding sites and target genes in maize.BMC Genomics. 2024 May 17;25(1):491. doi: 10.1186/s12864-024-10386-z. BMC Genomics. 2024. PMID: 38760741 Free PMC article. No abstract available.
Abstract
Background: Jasmonate (JA) is the important phytohormone to regulate plant growth and adaption to stress signals. MYC2, an bHLH transcription factor, is the master regulator of JA signaling. Although MYC2 in maize has been identified, its function remains to be clarified.
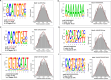
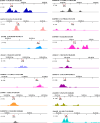
Results: To understand the function and regulatory mechanism of MYC2 in maize, the joint analysis of DAP-seq and RNA-seq is conducted to identify the binding sites and target genes of ZmMYC2. A total of 3183 genes are detected both in DAP-seq and RNA-seq data, potentially as the directly regulating genes of ZmMYC2. These genes are involved in various biological processes including plant growth and stress response. Besides the classic cis-elements like the G-box and E-box that are bound by MYC2, some new motifs are also revealed to be recognized by ZmMYC2, such as nGCATGCAnn, AAAAAAAA, CACGTGCGTGCG. The binding sites of many ZmMYC2 regulating genes are identified by IGV-sRNA.
Conclusions: All together, abundant target genes of ZmMYC2 are characterized with their binding sites, providing the basis to construct the regulatory network of ZmMYC2 and better understanding for JA signaling in maize.
Keywords: cis-element; Jasmonate; MYC2; Maize; Transcription factor.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

