Phenotypic and molecular characterization of β-lactamase-producing Klebsiella species among children discharged from hospital in Western Kenya
- PMID: 38654237
- PMCID: PMC11040804
- DOI: 10.1186/s12866-024-03284-7
Phenotypic and molecular characterization of β-lactamase-producing Klebsiella species among children discharged from hospital in Western Kenya
Abstract
Background: The emergence and spread of β-lactamase-producing Klebsiella spp. has been associated with a substantial healthcare burden resulting in therapeutic failures. We sought to describe the proportion of phenotypic resistance to commonly used antibiotics, characterize β-lactamase genes among isolates with antimicrobial resistance (AMR), and assess the correlates of phenotypic AMR in Klebsiella spp. isolated from stool or rectal swab samples collected from children being discharged from hospital.
Methods: We conducted a cross-sectional study involving 245 children aged 1-59 months who were being discharged from hospitals in western Kenya between June 2016 and November 2019. Whole stool or rectal swab samples were collected and Klebsiella spp. isolated by standard microbiological culture. β-lactamase genes were detected by PCR whilst phenotypic antimicrobial susceptibility was determined using the disc diffusion technique following standard microbiology protocols. Descriptive analyses were used to characterize phenotypic AMR and carriage of β-lactamase-producing genes. The modified Poisson regression models were used to assess correlates of phenotypic beta-lactam resistance.
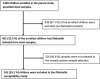
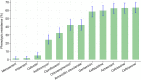
Results: The prevalence of β-lactamase carriage among Klebsiella spp. isolates at hospital discharge was 62.9% (154/245). Antibiotic use during hospitalization (adjusted prevalence ratio [aPR] = 4.51; 95%CI: 1.79-11.4, p < 0.001), longer duration of hospitalization (aPR = 1.42; 95%CI: 1.14-1.77, p < 0.002), and access to treated water (aPR = 1.38; 95%CI: 1.12-1.71, p < 0.003), were significant predictors of phenotypically determined β-lactamase. All the 154 β-lactamase-producing Klebsiella spp. isolates had at least one genetic marker of β-lactam/third-generation cephalosporin resistance. The most prevalent genes were blaCTX-M 142/154 (92.2%,) and blaSHV 142/154 (92.2%,) followed by blaTEM 88/154 (57.1%,) and blaOXA 48/154 (31.2%,) respectively.
Conclusion: Carriage of β-lactamase producing Klebsiella spp. in stool is common among children discharged from hospital in western Kenya and is associated with longer duration of hospitalization, antibiotic use, and access to treated water. The findings emphasize the need for continued monitoring of antimicrobial susceptibility patterns to inform the development and implementation of appropriate treatment guidelines. In addition, we recommend measures beyond antimicrobial stewardship and infection control within hospitals, improved sanitation, and access to safe drinking water to mitigate the spread of β-lactamase-producing Klebsiella pathogens in these and similar settings.
Keywords: Klebsiella spp; Antimicrobial resistance; Beta-lactams; Cephalosporins; Extended Spectrum Beta lactamases.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Antimicrobial resistance and prevalence of extended-spectrum beta-lactamase-producing Klebsiella species in East Tennessee dairy farms.Microbiol Spectr. 2024 Oct 3;12(10):e0353723. doi: 10.1128/spectrum.03537-23. Epub 2024 Sep 6. Microbiol Spectr. 2024. PMID: 39240080 Free PMC article.
-
Plasmid-mediated quinolone resistance genes detected in Ciprofloxacin non-susceptible Escherichia coli and Klebsiella isolated from children under five years at hospital discharge, Kenya.BMC Microbiol. 2023 May 13;23(1):129. doi: 10.1186/s12866-023-02849-2. BMC Microbiol. 2023. PMID: 37173674 Free PMC article.
-
Travel to Asia is a strong predictor for carriage of cephalosporin resistant E. coli and Klebsiella spp. but does not explain everything; prevalence study at a Norwegian hospital 2014-2016.Antimicrob Resist Infect Control. 2018 Nov 29;7:146. doi: 10.1186/s13756-018-0429-7. eCollection 2018. Antimicrob Resist Infect Control. 2018. PMID: 30534366 Free PMC article.
-
Antibiotic resistance in Indonesia: A systematic review and meta-analysis of extended-spectrum beta-lactamase-producing bacteria (2008-2024).Trop Med Int Health. 2025 Apr;30(4):246-259. doi: 10.1111/tmi.14090. Epub 2025 Jan 31. Trop Med Int Health. 2025. PMID: 39888153
-
From soil to surface water: exploring Klebsiella 's clonal lineages and antibiotic resistance odyssey in environmental health.BMC Microbiol. 2025 Feb 27;25(1):97. doi: 10.1186/s12866-025-03798-8. BMC Microbiol. 2025. PMID: 40012032 Free PMC article. Review.
Cited by
-
Neonatal Colonization With Antibiotic-Resistant Pathogens in Low- and Middle-Income Countries: A Systematic Review and Meta-Analysis.JAMA Netw Open. 2024 Nov 4;7(11):e2441596. doi: 10.1001/jamanetworkopen.2024.41596. JAMA Netw Open. 2024. PMID: 39499519 Free PMC article.
-
β-Lactamase and Macrolide Resistance Gene Carriage in Escherichia coli Isolates Among Children Discharged From Inpatient Care in Western Kenya: A Cross-sectional Study.Open Forum Infect Dis. 2024 Jun 3;11(6):ofae307. doi: 10.1093/ofid/ofae307. eCollection 2024 Jun. Open Forum Infect Dis. 2024. PMID: 38938894 Free PMC article.
-
Impact of Macrolide Resistance on Azithromycin for Prevention of Rehospitalization or Death Among Children Discharged From Hospitals in Western Kenya.J Infect Dis. 2025 Aug 14;232(2):e301-e308. doi: 10.1093/infdis/jiaf208. J Infect Dis. 2025. PMID: 40257829 Free PMC article. Clinical Trial.
References
-
- Godman B, Egwuenu A, Wesangula E, Schellack N, Kalungia AC, Tiroyakgosi C, et al. Expert Opinion on Drug Safety ISSN: (Print) (Online) Journal homepage: https://www.tandfonline.com/loi/ieds20 Tackling antimicrobial resistance across sub-Saharan Africa: current challenges and implications for the future. 2022. 10.1080/14740338.2022.2106368. Cited 2022 Nov 18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous