A protocol for annotation of total body photography for machine learning to analyze skin phenotype and lesion classification
- PMID: 38654834
- PMCID: PMC11035726
- DOI: 10.3389/fmed.2024.1380984
A protocol for annotation of total body photography for machine learning to analyze skin phenotype and lesion classification
Abstract
Introduction: Artificial Intelligence (AI) has proven effective in classifying skin cancers using dermoscopy images. In experimental settings, algorithms have outperformed expert dermatologists in classifying melanoma and keratinocyte cancers. However, clinical application is limited when algorithms are presented with 'untrained' or out-of-distribution lesion categories, often misclassifying benign lesions as malignant, or misclassifying malignant lesions as benign. Another limitation often raised is the lack of clinical context (e.g., medical history) used as input for the AI decision process. The increasing use of Total Body Photography (TBP) in clinical examinations presents new opportunities for AI to perform holistic analysis of the whole patient, rather than a single lesion. Currently there is a lack of existing literature or standards for image annotation of TBP, or on preserving patient privacy during the machine learning process.
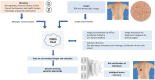
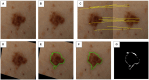
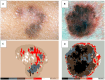
Methods: This protocol describes the methods for the acquisition of patient data, including TBP, medical history, and genetic risk factors, to create a comprehensive dataset for machine learning. 500 patients of various risk profiles will be recruited from two clinical sites (Australia and Spain), to undergo temporal total body imaging, complete surveys on sun behaviors and medical history, and provide a DNA sample. This patient-level metadata is applied to image datasets using DICOM labels. Anonymization and masking methods are applied to preserve patient privacy. A two-step annotation process is followed to label skin images for lesion detection and classification using deep learning models. Skin phenotype characteristics are extracted from images, including innate and facultative skin color, nevi distribution, and UV damage. Several algorithms will be developed relating to skin lesion detection, segmentation and classification, 3D mapping, change detection, and risk profiling. Simultaneously, explainable AI (XAI) methods will be incorporated to foster clinician and patient trust. Additionally, a publicly released dataset of anonymized annotated TBP images will be released for an international challenge to advance the development of new algorithms using this type of data.
Conclusion: The anticipated results from this protocol are validated AI-based tools to provide holistic risk assessment for individual lesions, and risk stratification of patients to assist clinicians in monitoring for skin cancer.
Keywords: artificial intelligence; computer image analyses; computer—aided diagnosis; dermatology; melanoma; total body photography.
Copyright © 2024 Primiero, Betz-Stablein, Ascott, D’Alessandro, Gaborit, Fricker, Goldsteen, González-Villà, Lee, Nazari, Nguyen, Ntouskos, Pahde, Pataki, Quintana, Puig, Rezze, Garcia, Soyer and Malvehy.
Conflict of interest statement
HS is a shareholder of MoleMap NZ Limited and e-derm consult GmbH and undertakes regular teledermatological reporting for both companies, Medical Consultant for Canfield Scientific Inc., Blaze Bioscience Inc., and a Medical Advisor for First Derm. JM and SP are co-founders of Diagnosis Dermatologica and Athena Care and investors in Dermavision. JM received honorarium from Canfield Scientific for consultancy and Fotofinder for educational activities. NA was employed by V7. BD was employed by Canfield Scientific Inc. SG was employed by ISAHIT. PF and HN were employed by Torus Actions & Belle.ai. AG was employed by IBM Research. SG-V and JQ were employed by Coronis Computing SL. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures

References
-
- Bohr A, Memarzadeh K. The rise of artificial intelligence in healthcare applications. Artif Intellig Healthcare. (2020):25–60. doi: 10.1016/B978-0-12-818438-7.00002-2 - DOI
-
- Kshatri SS, Singh D. Convolutional neural network in medical image analysis: a review. Arch Comput Methods Eng. (2023) 30:2793–810. doi: 10.1007/s11831-023-09898-w - DOI
LinkOut - more resources
Full Text Sources

