A bioinformatic approach to identify confirmed and probable CRISPR-Cas systems in the Acinetobacter calcoaceticus- Acinetobacter baumannii complex genomes
- PMID: 38655087
- PMCID: PMC11035748
- DOI: 10.3389/fmicb.2024.1335997
A bioinformatic approach to identify confirmed and probable CRISPR-Cas systems in the Acinetobacter calcoaceticus- Acinetobacter baumannii complex genomes
Abstract
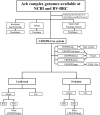
Introduction: The Acinetobacter calcoaceticus-Acinetobacter baumannii complex, or Acb complex, consists of six species: Acinetobacter baumannii, Acinetobacter calcoaceticus, Acinetobacter nosocomialis, Acinetobacter pittii, Acinetobacter seifertii, and Acinetobacter lactucae. A. baumannii is the most clinically significant of these species and is frequently related to healthcare-associated infections (HCAIs). Clustered regularly interspaced short palindromic repeat (CRISPR) arrays and associated genes (cas) constitute bacterial adaptive immune systems and function as variable genetic elements. This study aimed to conduct a genomic analysis of Acb complex genomes available in databases to describe and characterize CRISPR systems and cas genes.
Methods: Acb complex genomes available in the NCBI and BV-BRC databases, the identification and characterization of CRISPR-Cas systems were performed using CRISPRCasFinder, CRISPRminer, and CRISPRDetect. Sequence types (STs) were determined using the Oxford scheme and ribosomal multilocus sequence typing (rMLST). Prophages were identified using PHASTER and Prophage Hunter.
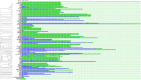
Results: A total of 293 genomes representing six Acb species exhibited CRISPR-related sequences. These genomes originate from various sources, including clinical specimens, animals, medical devices, and environmental samples. Sequence typing identified 145 ribosomal multilocus sequence types (rSTs). CRISPR-Cas systems were confirmed in 26.3% of the genomes, classified as subtypes I-Fa, I-Fb and I-Fv. Probable CRISPR arrays and cas genes associated with CRISPR-Cas subtypes III-A, I-B, and III-B were also detected. Some of the CRISPR-Cas systems are associated with genomic regions related to Cap4 proteins, and toxin-antitoxin systems. Moreover, prophage sequences were prevalent in 68.9% of the genomes. Analysis revealed a connection between these prophages and CRISPR-Cas systems, indicating an ongoing arms race between the bacteria and their bacteriophages. Furthermore, proteins associated with anti-CRISPR systems, such as AcrF11 and AcrF7, were identified in the A. baumannii and A. pittii genomes.
Discussion: This study elucidates CRISPR-Cas systems and defense mechanisms within the Acb complex, highlighting their diverse distribution and interactions with prophages and other genetic elements. This study also provides valuable insights into the evolution and adaptation of these microorganisms in various environments and clinical settings.
Keywords: Acinetobacter baumannii; Acinetobacter calcoaceticus–Acinetobacter baumannii complex; CRISPR systems; cas genes; prophages.
Copyright © 2024 Mancilla-Rojano, Flores, Cevallos, Ochoa, Parra-Flores, Arellano-Galindo, Xicohtencatl-Cortes and Cruz-Córdova.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Molecular Epidemiology of Acinetobacter calcoaceticus-Acinetobacter baumannii Complex Isolated From Children at the Hospital Infantil de México Federico Gómez.Front Microbiol. 2020 Oct 15;11:576673. doi: 10.3389/fmicb.2020.576673. eCollection 2020. Front Microbiol. 2020. PMID: 33178158 Free PMC article.
-
Acinetobacter calcoaceticus-baumannii complex prevalence, spatial-temporal distribution, and contamination sources in Canadian aquatic environments.Microbiol Spectr. 2024 Oct 3;12(10):e0150924. doi: 10.1128/spectrum.01509-24. Epub 2024 Sep 6. Microbiol Spectr. 2024. PMID: 39240108 Free PMC article.
-
The CRISPR-Cas system in clinical strains of Acinetobacter baumannii: an in-silico analysis.Lett Appl Microbiol. 2024 Jan 2;77(1):ovae003. doi: 10.1093/lambio/ovae003. Lett Appl Microbiol. 2024. PMID: 38211976
-
Advances in automated techniques to identify Acinetobacter calcoaceticus-Acinetobacter baumannii complex.Asian Biomed (Res Rev News). 2020 Oct 31;14(5):177-186. doi: 10.1515/abm-2020-0026. eCollection 2020 Oct. Asian Biomed (Res Rev News). 2020. PMID: 37551265 Free PMC article. Review.
-
Genotyping methods for monitoring the epidemic evolution of A. baumannii strains.J Infect Dev Ctries. 2015 Apr 15;9(4):347-54. doi: 10.3855/jidc.6201. J Infect Dev Ctries. 2015. PMID: 25881522 Review.
Cited by
-
Whole-genome sequencing of Acinetobacter baumannii clinical isolates from a tertiary hospital in Terengganu, Malaysia (2011-2020), revealed the predominance of the Global Clone 2 lineage.Microb Genom. 2025 Feb;11(2):001345. doi: 10.1099/mgen.0.001345. Microb Genom. 2025. PMID: 39908088 Free PMC article.
References
LinkOut - more resources
Full Text Sources

