An integrated analysis revealing the angiogenic function of TP53I11 in tumor microenvironment
- PMID: 38655349
- PMCID: PMC11036061
- DOI: 10.1016/j.heliyon.2024.e29504
An integrated analysis revealing the angiogenic function of TP53I11 in tumor microenvironment
Abstract
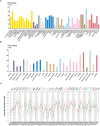
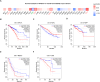
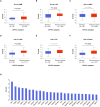
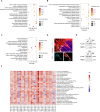
Despite growing evidence suggesting an important contribution of Tumor Protein P53 Inducible Protein 11 (TP53I11) in cancer progression, the role of TP53I11 remains unclear. Our first pan-cancer analysis of TP53I11 showed some tumor tissues displayed reduced TP53I11 expression compared to normal tissues, while others exhibited high TP53I11 expression. Meanwhile, TP53I11 expression carries a particular pan-cancer risk, as high TP53I11 expression levels are detrimental to survival for BRCA, KIRP, MESO, and UVM, but to beneficial survival for KIRC. We demonstrated that TP53I11 expression negatively correlates with DNA methylation in most cancers, and the S14 residue of TP53I11 is phosphorylated in several cancer types. Additionally, TP53I11 was found to be associated with endothelial cells in pan-cancer, and functional enrichment analysis provided strong evidence for its role in tumor angiogenesis. In vitro angiogenesis assays confirmed that TP53I11 can promote angiogenic function of human umbilical vein endothelial cells (HUVECs) in vitro. Mechanistic investigations reveal that TP53I11 is transcriptionally up-regulated by HIF2A under hypoxia.
Keywords: Angiogenesis; Endothelial cells; Immune infiltration; Prognosis; TP53I11.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


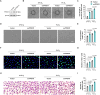
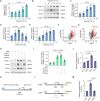
References
-
- Collins F.S., McKusick V.A. Implications of the human genome project for medical science. JAMA. 2001;285:540–544. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

